Pecinka Lab:Research: Difference between revisions
From OpenWetWare
Jump to navigationJump to search
Ales Pecinka (talk | contribs) No edit summary |
Ales Pecinka (talk | contribs) No edit summary |
||
| Line 29: | Line 29: | ||
<h3><font style="color:#F8B603;">Current Projects</font></h3> | <h3><font style="color:#F8B603;">Current Projects</font></h3> | ||
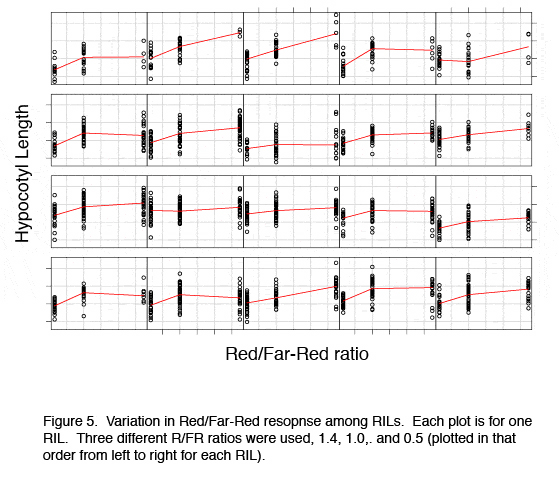
====Natural variation in | ====Natural variation in response to DNA replication stress==== | ||
We are interested to identify genes or natural alleles that confer an altered DNA repair capacity and are involved in fine-tuning of the DNA repair machinery under natural conditions. Currently, two QTL mapping projects are running in the lab focusing on responses on A. thaliana to UV-B light and DNA replication blocking agent hydroxyurea. | We are interested to identify genes or natural alleles that confer an altered DNA repair capacity and are involved in fine-tuning of the DNA repair machinery under natural conditions. Currently, two QTL mapping projects are running in the lab focusing on responses on A. thaliana to UV-B light and DNA replication blocking agent hydroxyurea. | ||
[[Image:Research_fig_5.jpg | center | Figure 5]] | [[Image:Research_fig_5.jpg | center | Figure 5]] | ||
| Line 39: | Line 38: | ||
xxxxxx | xxxxxx | ||
==== | ====Analysis of natural variation in response to UV-B light and genome-wide UV-B mutagenic effects==== | ||
An effective source of DNA damage in nature is UV light. To reduce other than DNA damage effects, we use UV-C radiation that is the most potent inducer of pyrimidine dimers - a specific type of crosslinks between two pyrimidine bases (C and T). Pyrimidine dimers cause a physical barrier for transcription and replication and therefore need to be repaired. This is facilitated by Arabidopsis photolyases UVR2 (PHR1) and UVR3. However, it is not clear how the whole machinery is regulated under natural latitudinal and altitudinal light intensity (including UV) gradients. To address this, we use different Arabidopsis natural accessions and screen for their relative survival after high dose of UV-C (Figure 1A). Analysis of approximately 100 accessions revealed great differences ranging from full resistance to full sensitivity (Figure 1B). | |||
====The role of epigenetic control of repetitive DNA in the genome maintenance==== | |||
A large part of eukaryotic genome consists of repetitive DNA. Repeats have important functions in e.g. constituting centromeres, telomeres and rDNAs and developmental regulation of gene expression (FWA REF). In addition, they contribute to genome instability and chromosomal rearrangements due to transposition activity and as frequent targets of homologous recombination machinery. To preserve genome integrity, repeat activity is suppressed by transcriptional gene silencing (TGS), an epigenetic control mechanism that brings DNA into a less accessible "heterochromatic" state via small interfering RNA directed deposition of silencing chromatin marks DNA methylation and histone H3 lysine 9 di-methylation. Our aim is to explore variability in control of TGS in the natural variants of A. thaliana, its close relatives and other Brassicaceae. | |||
We have identified A. thaliana accession that shows activation of TRANSCRIPTIONALLY SILENT INFORMATION (TSI; Steimer et al., 2001) – an ATHILA related retrotransposon (Figure 3). | |||
[[Image:Research_fig_5.jpg | center | Figure 5]] | |||
Several Arabidopsis mutants showing transcriptional activation of this element, e.g decreased dna methylation 1 (ddm1) and dna methyltransferase 1 (met1), are severely affected in their life-span and fitness (Kakutani et al., 1996; Mathieu et al., 2007). We are therefore interested to identify causal polymorphisms and to test their influence on the survival. Genetic mapping revealed one recessive and one dominant QTL in one genome and a recessive QTL with an additive effect on the TSI transcript amount in the other parental genome. Further mapping is in progress. | |||
The knowledge on TGS in plants is biased towards A. thaliana model system. However, A. thaliana has greatly reduced genome with only about 13% of repetitive DNA (Arabidopsis Genome Initiative, 2000) and therefore is rather an exception in the plant kingdom. We are interested in understanding TGS and epigenetic genome regulation also in other Brassicaceae with larger genomes and higher content of repetitive DNA. | |||
= | A. thaliana and its close relatives show largely uniform pattern of genome-wide repetive DNA distribution with majority of repeats being clustered in pericentromeric regions and repeat depleted chromosome arms. Recent study revealed an unusual distribution of heterochromatin in ''Ballantinia antipoda'' (2n = 12), an Australian ''Brassicaceae'' species closely related to ''A. thaliana'' (Mandakova et al., 2010). In addition to the typical pericentromeric heterochromatic blocks present in ''A. thaliana'', ''B. antipoda'' contains six large heterochromatic segments (HSs) covering 30 to 100% of a chromosome arm (Figure 4A). Another striking phenotype of HSs challenging current model of heterochromatin establishement derived from ''A. thaliana'' is an absence of DNA methylation (Figure 4B). Hybridization of ''B. antipoda'' chromosomes with a fluorescently labeled ''A. thaliana'' derived probes suggests that HSs are not consisiting of telomeric or rDNA sequences and do contain intersperesed single copy genic sequences. Although heterochromatin typically constitutes of repetitive DNA that is heavily DNA methylated, HSs of B. antipoda contain at least partially unique sequences, do not overlap with rDNAs and telomeric repeats and seem to be free of DNA methylation (Figure 4B and Mandakova et al., 2010). | ||
Currently, the principal sequence of HSs in B. antipoda is isolated and will be characterized with respect to DNA methylation, histone modifications and transcriptional activation pattern during different developmental stages and stress situations. | |||
<h3><font style="color:#F8B603;">Bibliography</font></h3> | <h3><font style="color:#F8B603;">Bibliography</font></h3> | ||
Revision as of 01:32, 5 June 2012
|
Centre of Plant Structural and Functional Genomics |