Pecinka Lab:Research: Difference between revisions
From OpenWetWare
Jump to navigationJump to search
Ales Pecinka (talk | contribs) No edit summary |
Ales Pecinka (talk | contribs) No edit summary |
||
| Line 7: | Line 7: | ||
Deoxyribonucleic acid (DNA) is the main genetic information storage molecule of all organisms and therefore it needs to be protected from sequence changes. This is ensured by genome maintenance mechanisms - proofreading activity of DNA polymerases, DNA repair and control of repetitive DNA sequences by epigenetic means. Defective or incomplete function of these mechanisms may result in rapid accumulation of mutations, chromosomal breakage or rearrangements, reduced fitness and development of diseases. | Deoxyribonucleic acid (DNA) is the main genetic information storage molecule of all organisms and therefore it needs to be protected from sequence changes. This is ensured by genome maintenance mechanisms - proofreading activity of DNA polymerases, DNA repair and control of repetitive DNA sequences by epigenetic means. Defective or incomplete function of these mechanisms may result in rapid accumulation of mutations, chromosomal breakage or rearrangements, reduced fitness and development of diseases. | ||
[[Image:Research_fig_1.jpg | center | Figure 1]] | [[Image:Research_fig_1.jpg | center | Figure 1]] | ||
| Line 27: | Line 29: | ||
<h3><font style="color:#F8B603;">Current Projects</font></h3> | <h3><font style="color:#F8B603;">Current Projects</font></h3> | ||
==== | ====Natural variation in the DNA repair mechanisms==== | ||
We are interested to identify genes or natural alleles that confer an altered DNA repair capacity and are involved in fine-tuning of the DNA repair machinery under natural conditions. Currently, two QTL mapping projects are running in the lab focusing on responses on A. thaliana to UV-B light and DNA replication blocking agent hydroxyurea. | |||
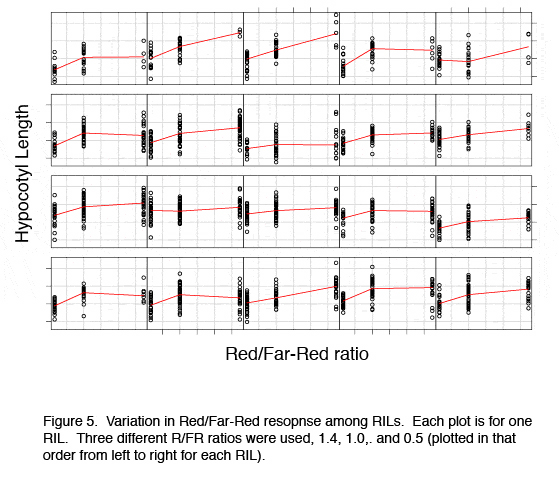
An effective source of DNA damage in nature is UV light. To reduce other than DNA damage effects, we use UV-C radiation that is the most potent inducer of pyrimidine dimers - a specific type of crosslinks between two pyrimidine bases (C and T). Pyrimidine dimers cause a physical barrier for transcription and replication and therefore need to be repaired. This is facilitated by Arabidopsis photolyases UVR2 (PHR1) and UVR3. However, it is not clear how the whole machinery is regulated under natural latitudinal and altitudinal light intensity (including UV) gradients. To address this, we use different Arabidopsis natural accessions and screen for their relative survival after high dose of UV-C (Figure 1A). Analysis of approximately 100 accessions revealed great differences ranging from full resistance to full sensitivity (Figure 1B). | |||
[[Image:Research_fig_5.jpg | center | Figure 5]] | [[Image:Research_fig_5.jpg | center | Figure 5]] | ||
Revision as of 14:15, 4 June 2012
|
Centre of Plant Structural and Functional Genomics |