PCR inhibitors: Difference between revisions
No edit summary |
No edit summary |
||
| (5 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
Many compounds also contained in the PCR template beside the target DNA can interfere with the polymerisation of nucleotides by the Taq or a related polymerase. This may result in false negative results, i.e. no PCR product even though the template is present. | Many compounds also contained in the PCR template beside the target DNA can interfere with the polymerisation of nucleotides by the Taq or a related polymerase. This may result in false negative results, i.e. no PCR product even though the template is present. | ||
== Mechanisms of inhibition == | |||
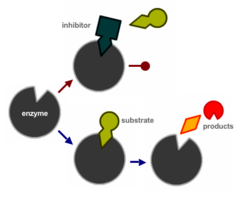
A multitude of mechanisms can cause a [[PCR]] to fail. Binding of an inhibitor to the active site of a DNA polymerase is schematically depicted on the right. Other reasons include sequestration of essentials co-factors like Mg<sup>2+</sup>, e.g. by chelators like [[EDTA]] used in [[TE buffer]]. Many PCR also fail due to difficult template sequences, for example GC-rich, that cause the polymerisation process to be inefficient or even fail. In many cases, several factor only decrease the efficiency of the PCR but together lead the PCR product falling below the detection threshold. | |||
== Inhibitors from the purification process == | == Inhibitors from the purification process == | ||
| Line 21: | Line 25: | ||
== Template inhibition == | == Template inhibition == | ||
The amount of template and the ratio of template to primer is an important parameter to optimise for difficult PCRs. Adding more template can in some cases decrease the amount of PCR product obtained. However, since inhibitors often enter the PCR reaction via the template preparation, decreasing template volume may improve PCR results indirectly by reducing the quantity of inhibiting contaminations. | |||
Typical starting amounts for genomic DNA template are 25-1000 ng in a 25 µl PCR reaction. Starting amounts for plasmid templates are generally much lower at 5-50ng per 25µl PCR reaction. | |||
== See also == | == See also == | ||
* [[ | * [[Image:2stars.png]] [http://www.med.yale.edu/genetics/ward/tavi/Trblesht.html PCR trouble shooting guide by O. Henegariu from Yale] | ||
* [[Maheshri:PCR]] - has some tips on how to avoid inhibition | * [[Image:2stars.png]] [http://www.neb.com/nebecomm/products/faqproductM0273.asp PCR trouble shooting guide by NEB] | ||
* [[Image:1star.png]] [http://en.wikipedia.org/wiki/Polymerase_chain_reaction_inhibitors PCR inhibitor at the Wikipedia] | |||
* [[Image:1star.png]] [[Maheshri:PCR]] - has some tips on how to avoid inhibition | |||
* [[Image:1star.png]] [http://www.embl-hamburg.de/~geerlof/webPP/genetoprotein/clo_pcr_strategy/optimizing_PCR.html PCR optimization guide by A Geerlof, EMBL Hamburg] | |||
<small>(stars indicate relevance of the links)</small> | |||
* [[PCR]] - general, simple overview of polymerase chain reaction | * [[PCR]] - general, simple overview of polymerase chain reaction | ||
* [[PCR techniques]] - hub page for PCR-based techniques | * [[PCR techniques]] - hub page for PCR-based techniques | ||
* [[Purification of DNA]] | * [[Purification of DNA]] | ||
[[Category:PCR]] [[Category:DNA]] [[Category:In vitro]] [[Category:Chemicals]] | [[Category:PCR]] [[Category:DNA]] [[Category:In vitro]] [[Category:Chemicals]] | ||
Latest revision as of 05:57, 7 July 2010
Many compounds also contained in the PCR template beside the target DNA can interfere with the polymerisation of nucleotides by the Taq or a related polymerase. This may result in false negative results, i.e. no PCR product even though the template is present.
Mechanisms of inhibition
A multitude of mechanisms can cause a PCR to fail. Binding of an inhibitor to the active site of a DNA polymerase is schematically depicted on the right. Other reasons include sequestration of essentials co-factors like Mg2+, e.g. by chelators like EDTA used in TE buffer. Many PCR also fail due to difficult template sequences, for example GC-rich, that cause the polymerisation process to be inefficient or even fail. In many cases, several factor only decrease the efficiency of the PCR but together lead the PCR product falling below the detection threshold.
Inhibitors from the purification process
- alcohols like ethanol and 2-propanol (isopropanol) from DNA template precipitation (Qiagen test of sequencing inhibition by Welters et al. 1997)
- organic solvents like phenol from phenol/chloroform purification (PMID 8136148)
- salts like KCl, NaCl from precipitation
- detergents like SDS,.. from membrane lysis (PMID 2223070)
Inhibitors from the source tissue
- components of blood: heme, hemoglobin, lactoferrin, immunoglobin G (IgG)
- liver, digestive tract, feces: bile salts, polysaccharides
- connective tissue, skin: collagen, melanin
- urine: urea
See Rådström et al. 2004 (PMID 14764939) for a review on how to avoid inhibition from tissue components.
Template inhibition
The amount of template and the ratio of template to primer is an important parameter to optimise for difficult PCRs. Adding more template can in some cases decrease the amount of PCR product obtained. However, since inhibitors often enter the PCR reaction via the template preparation, decreasing template volume may improve PCR results indirectly by reducing the quantity of inhibiting contaminations.
Typical starting amounts for genomic DNA template are 25-1000 ng in a 25 µl PCR reaction. Starting amounts for plasmid templates are generally much lower at 5-50ng per 25µl PCR reaction.
See also
 PCR trouble shooting guide by O. Henegariu from Yale
PCR trouble shooting guide by O. Henegariu from Yale PCR trouble shooting guide by NEB
PCR trouble shooting guide by NEB PCR inhibitor at the Wikipedia
PCR inhibitor at the Wikipedia Maheshri:PCR - has some tips on how to avoid inhibition
Maheshri:PCR - has some tips on how to avoid inhibition PCR optimization guide by A Geerlof, EMBL Hamburg
PCR optimization guide by A Geerlof, EMBL Hamburg
(stars indicate relevance of the links)
- PCR - general, simple overview of polymerase chain reaction
- PCR techniques - hub page for PCR-based techniques
- Purification of DNA