OpenSourceMalaria:Triazolopyrazine (TP) Series: Difference between revisions
Matthew Todd (talk | contribs) |
Matthew Todd (talk | contribs) (note that this wiki is inactive) |
||
| (102 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
{{OpenSourceMalaria}} | {{OpenSourceMalaria}} | ||
'''Important Note - this page is no longer being updated. The active wiki is [https://github.com/OpenSourceMalaria/Series4/wiki here].''' | |||
=Open Source Malaria Series 4: The Triazolopyrazine (TP) Series= | =Open Source Malaria Series 4: The Triazolopyrazine (TP) Series= | ||
== | ==Resources== | ||
'''Molecules and How to Visualize Them''' | |||
<br> | |||
[http://www.openmolecules.org/datawarrior/download.html OSIRIS DataWarrior] (general purpose open-source chemoinformatics software) <br> | |||
[http://tinyurl.com/OSM-Compounds Google Sheet of all OSM Compounds] (Can filter Series 4).<br> | |||
[http://tinyurl.com/OSM-Assays Sister sheet that explains column headings (i.e. assays used)].<br> | |||
[http://www.cheminfo.org/flavor/malaria/Display_data.html Visualisation of OSM molecules on Cheminfo]. The data may be visualized with [http://macinchem.org/reviews/vortex/tut26/scripting_vortex26.php Vortex]. The Series 4 SDF can be generated on Cheminfo - [http://malaria.ourexperiment.org/data/files/17162/malaria-2016-05-18.sdf here] is a downloadable version from May 2016. | |||
'''Lab Notebooks''' | |||
[https:// | [http://malaria.ourexperiment.org/triazolopyrazine_se The principal Labtrove ELN]<br> | ||
[https://au-mynotebook.labarchives.com/share/Edwin%2520Tse/MTYzLjh8MTUwOS8xMjYvVHJlZU5vZGUvMjUxNDEwNDEwOHw0MTUuOA== Ed Tse's Labarchives ELN]<br> | |||
[http://tinyurl.com/OSM-Jasper Jasper Tyler's Lab Notebook]<br> | |||
[https://mynotebook.labarchives.com/share/MCPHS%2520MedChem/MzUxLjB8MzY3Ny8yNzAvVHJlZU5vZGUvNDA0ODg3Njc4OHw4OTEuMA== Chase Smith's Labarchives ELN]<br> | |||
[http://malaria.ourexperiment.org/data/files/7384/FINAL%20REPORT%20MMV670652.doc Old document from CRO describing synthetic chemistry of some members of series]<br> | |||
'''Papers''' | |||
First paper based on this series is [https://github.com/OpenSourceMalaria/OSMSeries4Paper1 being written up here]. | |||
[http://www.nature.com/nrd/journal/v15/n10/full/nrd.2016.154.html Relevant 2016 review from MMV]. [https://malariajournal.biomedcentral.com/articles/10.1186/s12936-016-1675-x 2017 New developments in anti-malarial target candidate and product profiles] | |||
'''How To Join In''' | |||
[https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues?labels=Series+4&milestone=&page=1&state=open The To Do List]<br> | |||
How to involve yourself in OSM is described in the [http://opensourcemalaria.org/ Landing Page] under "Join the Team". | |||
'''Document Archive''' | |||
[http://malaria.ourexperiment.org/uri/404# Initial briefing document on the series written for MMV] and a [http://malaria.ourexperiment.org/uri/406 PDF summary of pharmacokinetics and efficacy]; largely folded into the page below. (A minor error in the briefing document referring to the amides has been [http://malaria.ourexperiment.org/uri/42d clarified].) | |||
[http://malaria.ourexperiment.org/biological_data/10041 PDF of the Compounds Originally Inherited from MMV] including structures and potencies ([http://malaria.ourexperiment.org/osdd_malaria_shared/8106/MMV_triazolopyrazine_data.html | |||
older version of this file]) | |||
[http://malaria.ourexperiment.org/osdd_malaria_shared/8292/Collected_Amides_in_the_TP_Series_Triazolopyrazines_Series_4.html Old Summary of Data on Amides in the Series]<br> | |||
[http://malaria.ourexperiment.org/osdd_malaria_shared/8585 ELN post containing many of the raw Chemdraw and picture files for this wiki page] - if you generate new pictures, add to the ELN page. This resource needs porting to Github. | |||
Project Reports: [http://malaria.ourexperiment.org/osdd_malaria_shared/11335 Devon Scott and Eduvie Omene, University of Edinburgh], [https://figshare.com/articles/Developments_of_Routes_to_Access_and_Functionalise_1_2_4_Triazolo_4_3_a_pyrazine_Amides_as_New_Antimalarials/1301073/1 Tom MacDonald, The University of Sydney]. | |||
'''Online Meetings''' | |||
[https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/386 Most recent online meeting (May 2016) relevant to this series]. [https://www.dropbox.com/s/9643p9d7mer8vfa/2016-05-24%2013.02%20Malaria%20open%20source%20drug%20discovery%20design%20meeting.mp4?dl=0 Recording]. | |||
This page is at http://tinyurl.com/OSM-Series4 | |||
==Introduction== | |||
== | ===Preamble=== | ||
The Triazolopyrazine (TP) Series, or ''Series 4'', is the latest of the OSM series. It was announced by [http://www.mmv.org/newsroom/news/potential-new-class-antimalarials-now-open-source MMV] and on the OSM blog (''via'' [http://malaria.ourexperiment.org/osdd_malaria_shared/7949/A_New_Triazolopyrazine_Series_for_OSM__Series_4.html the briefing document] and as [http://malaria.ourexperiment.org/the_osm_blog/7954/Announcing_OSM_Series_4__the_Triazolopyrazines.html a general description]) on September 10th 2013. | |||
[[Image:Representative TP Compound.png|thumb|center|250px|A Representative Series 4 Triazolopyrazine]] | |||
The series arises from work performed at Pfizer (that cannot be fully disclosed) which was followed by some hit-to-lead work funded directly by MMV and performed by a CRO (which can). The series includes many potent compounds, some with promising physicochemical properties, that are non-toxic and which clear assays such as hERG. Most importantly, members of the series have proven to be potent ''in vivo''. | |||
==Current Series Aims== | |||
1. Lead optimisation, to improve solubility and metabolic stability while maintaining potency.<br> | |||
2. Validation of PfATP4 activity as mechanism of action ''via'' experimental and computational means.<br> | |||
3. To source more chemical and biological inputs from the widest possible community.<br> | |||
==Notable Points about Series | ==Notable Points about The Series== | ||
* Compounds in this series have been identified down to 16 nM potency. | * Compounds in this series have been identified down to 16 nM potency. | ||
* Seems to have good ''in vitro'' HLM and hHEP stability CL<sub>int</sub> < 8.1 is compatible with 10 nM potency. | * Seems to have good ''in vitro'' HLM and hHEP stability: CL<sub>int</sub> < 8.1 is compatible with 10 nM potency. | ||
* RLM remains stubbornly high, particularly for the more potent analogues translating to short half-lives in rat PK. It's possible that rat metabolism may not be a good model for human metabolism for this series. | * RLM remains stubbornly high, particularly for the more potent analogues translating to short half-lives in rat PK. It's possible that rat metabolism may not be a good model for human metabolism for this series. | ||
* Series appears to have little polypharmacology or cytotoxicity. | * Series appears to have little polypharmacology or cytotoxicity. | ||
* There is strong correlation between activity ''vs.'' Pfal and activity in Kiaran Kirk’s ion regulation assay, implying the mechanism of action is inhibition of PfATP4. | * There is strong correlation between activity ''vs.'' Pfal and activity in Kiaran Kirk’s ion regulation assay, implying the mechanism of action is inhibition of PfATP4. | ||
==Concerns about | ==Concerns about The Series== | ||
* Although [http://en.wikipedia.org/wiki/Dofetilide# dofetilide] binding looks weak or nil, the series has shown activity in a patch clamp assay at Essen (1-10 μM) which is quite potent though with a window of >100 fold over Pfal potency. | * Although [http://en.wikipedia.org/wiki/Dofetilide# dofetilide] binding looks weak or nil, the series has shown activity in a patch clamp assay at Essen (1-10 μM) which is quite potent though with a window of >100 fold over Pfal potency. | ||
| Line 94: | Line 97: | ||
[[Image:Core Mods Triazole.png|thumb|center|600px|Modification of Core Triazole]] | [[Image:Core Mods Triazole.png|thumb|center|600px|Modification of Core Triazole]] | ||
The ring system in MMV669846 (imidazo[1,2-a]pyrazine) is also contained in the similar-looking Novartis series around compound [http://aac.asm.org/content/early/2013/12/17/AAC.01927-13.abstract KAI407], but the compounds [http://malaria.ourexperiment.org/triazolopyrazine_se/8912 apparently possess distinct mechanisms of action]. | |||
Modifications to the pyrazine part of the core were not well tolerated. Several aromatic variations were tried. | Modifications to the pyrazine part of the core were not well tolerated. Several aromatic variations were tried. | ||
| Line 104: | Line 109: | ||
As most of the analogs arising from core modifications were >1 μM potency, fewer were tested in RLM (though quite a few were tested in HLM). Of the 4 tested in RLM, the greatest stability had a Clint of 109, (HLM 9.5), several had HLM Clint 8 or less, particularly after moving or removing the N from the pyrazine ring. ''(<--need to locate primary data, since this disagrees with what is in the pictures here - MHT)'' | As most of the analogs arising from core modifications were >1 μM potency, fewer were tested in RLM (though quite a few were tested in HLM). Of the 4 tested in RLM, the greatest stability had a Clint of 109, (HLM 9.5), several had HLM Clint 8 or less, particularly after moving or removing the N from the pyrazine ring. ''(<--need to locate primary data, since this disagrees with what is in the pictures here - MHT)'' | ||
A selection of triazolopyridines were synthesised by Jamie Scott at The University of Sydney. Both ethers and amides were synthesised and found to be less potent than the corresponding trizaolopyrazine compounds. | |||
[[Image:Core Mod Triazolopyridines.png|thumb|center|800px|Modification of Core Pyrazine, Replacing with a Pyridine]] | |||
===Modification of Pyrazine Substitution Pattern=== | ===Modification of Pyrazine Substitution Pattern=== | ||
| Line 128: | Line 137: | ||
[[Image:CH3 vs CHF2 vs CF3 Northeast.png|thumb|center|500px|Sensitivity to Methoxy Substitution of the Triazole's Aromatic Substituent]] | [[Image:CH3 vs CHF2 vs CF3 Northeast.png|thumb|center|500px|Sensitivity to Methoxy Substitution of the Triazole's Aromatic Substituent]] | ||
A selection of benzylic alcohols were synthesised containing different substitution on the triazole ring. Once again, a paraphenyl-OCHF<sub>2</sub> group led to potency whereas interestingly, a phenyl substituent led to inactivity, 2-substituted pyridine displayed micromolar activity, again suggesting that substituted pyridines could be further explored. Both cyclic and acyclic aliphatic compounds were inactive. | |||
[[Image:Benzylic Alcohols.png|thumb|center|600px|Sensitivity to different aromatic and aliphatic substituents]] | |||
===Pyrazine Side Chain Modifications - Ethers=== | ===Pyrazine Side Chain Modifications - Ethers=== | ||
| Line 138: | Line 151: | ||
* Constraining the linear side-chain into ring systems (e.g, azetidines, pyrrolidines, pyrazoles) severely reduced potency. | * Constraining the linear side-chain into ring systems (e.g, azetidines, pyrrolidines, pyrazoles) severely reduced potency. | ||
[[Image: | [[Image:The Ether Library June15 update.png|thumb|center|850px|Ethers Synthesised and Tested pre-OSM]] | ||
A 2-naphthol substituent on the pyrazine ring showed reasonable potency against Pfal (IC50 114 nM) but suffered from poor RLM stability (''what is the number?''). Hetero-analogs of 2-naphthol e.g. indole, indazole, quinoline, chroman, benzisoxazole, quinazoline, exhibited reduced potency. | A 2-naphthol substituent on the pyrazine ring showed reasonable potency against Pfal (IC50 114 nM) but suffered from poor RLM stability (''what is the number?''). Hetero-analogs of 2-naphthol e.g. indole, indazole, quinoline, chroman, benzisoxazole, quinazoline, exhibited reduced potency. | ||
| Line 144: | Line 157: | ||
In a separate attempt to mitigate the potential metabolism in the ethyl chain it was replaced by an aromatic group through inclusion of a phenol substituent on the pyrazine ring. Some substituted phenolates were metabolically more stable '''in vitro''' as well as ''in vivo'' in rat, although with reduced potency. (''what are the numbers?'') | In a separate attempt to mitigate the potential metabolism in the ethyl chain it was replaced by an aromatic group through inclusion of a phenol substituent on the pyrazine ring. Some substituted phenolates were metabolically more stable '''in vitro''' as well as ''in vivo'' in rat, although with reduced potency. (''what are the numbers?'') | ||
Considering that the benzylic position in the phenethyl side-chain is prone to metabolic oxidation, several compounds having mono- and di-substitution in the benzylic position were made. Di-substitution lowered the potency considerably whereas mono-substitution with OMe, OCHF<sub>2</sub>, CH<sub>2</sub>OH, NMe<sub>2</sub> groups retained good potency. Additional substitution alpha- to the ether oxygen led to complete loss of potency. The alpha-OCHF<sub>2</sub> compound MMV670652, with a p-CN-phenyl group on the triazole ring, showed better RLM stability (cLogP effect). (''what is the number?'' | Considering that the benzylic position in the phenethyl side-chain is prone to metabolic oxidation, several compounds having mono- and di-substitution in the benzylic position were made. Di-substitution lowered the potency considerably whereas mono-substitution with OMe, OCHF<sub>2</sub>, CH<sub>2</sub>OH, NMe<sub>2</sub> groups retained good potency. This pattern was confirmed with the synthesis of phenyl analogues (MMV6888-96, -98 and 99). Synthesis of the corresponding dimethylamine is ongoing. Additional substitution alpha- to the ether oxygen led to complete loss of potency. The alpha-OCHF<sub>2</sub> compound MMV670652, with a p-CN-phenyl group on the triazole ring, showed better RLM stability (cLogP effect). (''what is the number?'' | ||
Chris Swain suggested the synthesis of some benzyl ethers. MMV675959 was found to possess moderate activity. More of these compounds may be synthesised in July/Aug 2015, pending online consultation. In particular, 2-, 3- and 4-chlorosubstituted benzene and alpha-substituted benzylic ethers are of interest. | |||
[[Image:Benzylic Ethers.png|thumb|center|150px|Ethers Synthesised and Tested pre-OSM]] | |||
Planning of ether analogs: GHI [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/174 174] and [http://malaria.ourexperiment.org/the_osm_blog/9601 blog appeal]. Planning of more analogs in July 2014: GHI [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/232 232]. | Planning of ether analogs: GHI [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/174 174] and [http://malaria.ourexperiment.org/the_osm_blog/9601 blog appeal]. Planning of more analogs in July 2014: GHI [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/232 232]. | ||
| Line 163: | Line 180: | ||
[[Image:The Other Linkers Abbreviated.png|thumb|center|800px|''N''- and ''C''-Based Attachment of Side Chains to Pyrazine]] | [[Image:The Other Linkers Abbreviated.png|thumb|center|800px|''N''- and ''C''-Based Attachment of Side Chains to Pyrazine]] | ||
===Vito Spadavecchio's potency predictor=== | |||
This is a Linux based tool that uses regression to predict the potency of a arbitrary series 4 molecule: https://github.com/spadavec/OSM-ML- | |||
==Physicochem/metabolism/PK== | ==Physicochem/metabolism/PK== | ||
The scheme below shows all the inherited met/PK data inherited, plus additional [http://malaria.ourexperiment.org/uri/490 data] for compounds more recently [http://malaria.ourexperiment.org/uri/457 sent] to Monash (green). (Associated discussion: GHI [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/107 107]). | The scheme below shows all the inherited met/PK data inherited, plus additional [http://malaria.ourexperiment.org/uri/490 data] for compounds more recently [http://malaria.ourexperiment.org/uri/457 sent] to Monash (green). (Associated discussion: GHI [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/107 107]). [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/471 Dec 2016: Tool for predicting sites of metabolism] | ||
[[Image:Metabolism Summary.png|thumb|center|900px|Series 4 Compounds Evaluated for Solubility and Metabolic Clearance]] | [[Image:Metabolism Summary.png|thumb|center|900px|Series 4 Compounds Evaluated for Solubility and Metabolic Clearance]] | ||
For the more recent set, solubility was still seen as low and needs improving. The metabolic stability measured for the recent set appears not to be a mouse-specific event and needs to be improved to deliver a candidate. Met ID data (below) could help in the design of new analogs with improved stability. Is there a correlation between metabolic stability and Log D? | For the more recent set, solubility was still seen as low and needs improving. The metabolic stability measured for the recent set appears not to be a mouse-specific event and needs to be improved to deliver a candidate. Met ID data (below) could help in the design of new analogs with improved stability. Is there a correlation between metabolic stability and Log D? | ||
Some [http://malaria.ourexperiment.org/biological_data/7934 inherited data] where the nature of the assay used was not clear: MMV669360 CLint 142 uL/min/kg and t1/2 = 12 min. | |||
The PK curve for the relatively weak amide MMV670246 is shown below for oral & IV legs & parameters. The original data are contained in [http://openwetware.org/wiki/Image:MMV670246_rat_PK_data.png this picture]. | The PK curve for the relatively weak amide MMV670246 is shown below for oral & IV legs & parameters. The original data are contained in [http://openwetware.org/wiki/Image:MMV670246_rat_PK_data.png this picture]. | ||
| Line 197: | Line 219: | ||
[[Image:AO Results from Pfizer.png|thumb|center|600px|Aldehyde Oxidase Assay Results from Pfizer]] | [[Image:AO Results from Pfizer.png|thumb|center|600px|Aldehyde Oxidase Assay Results from Pfizer]] | ||
For reference there is a [http://www.cambridgemedchemconsulting.com/resources/ADME/aldehyde_oxidase.html very nice summary of AO] over at Cambridge MedChem Consulting | |||
==Liver Stage== | |||
Two compounds have (MMV669844 and MMV670944, below) been evaluated ''vs.'' Pb liver schizonts and found to possess low activity ''vs.'' their blood stage potencies. Data [http://malaria.ourexperiment.org/triazolopyrazine_se/8912/Similarity_of_Series_4_Compounds_to_KAI407.html here] (along with a discussion of the similarity of Series 4 to the Novartis compound KAI407). (The [http://malaria.ourexperiment.org/osdd_malaria_shared/7949/post.html original Series 4 briefing document] stated the series had "no activity against Winzeler’s Pb liver stage" but there were no associated data to support this statement.) | |||
[[Image:Liver Stage Data.png|thumb|center|400px|Liver Stage Activities of Two Series 4 Compounds]] | |||
==Gametocyte Stage== | |||
The [http://malaria.ourexperiment.org/osdd_malaria_shared/7949/post.html original Series 4 briefing document] stated the series had "no or weak >>1uM activity against gametocytes" but there are no associated data to support this statement. | |||
==In Vivo Efficacy== | ==In Vivo Efficacy== | ||
| Line 224: | Line 258: | ||
The following five compounds were evaluated in parasite ion regulation assays in the [http://biology.anu.edu.au/kiaran_kirk/ Kirk Laboratory]; the hypothesis is that PfATP4 is a Na<sup>+</sup> ATPase that exports Na<sup>+</sup> and imports H<sup>+</sup> (or equivalent) and that the effects of the compounds on Na<sup>+</sup> concentration and pH are attributable to inhibition of this activity. Structures, potency, metabolism/solubility and raw PfATP4 assay data are [http://malaria.ourexperiment.org/biological_data/7934/ here]. | The following five compounds were evaluated in parasite ion regulation assays in the [http://biology.anu.edu.au/kiaran_kirk/ Kirk Laboratory]; the hypothesis is that PfATP4 is a Na<sup>+</sup> ATPase that exports Na<sup>+</sup> and imports H<sup>+</sup> (or equivalent) and that the effects of the compounds on Na<sup>+</sup> concentration and pH are attributable to inhibition of this activity. Structures, potency, metabolism/solubility and raw PfATP4 assay data are [http://malaria.ourexperiment.org/biological_data/7934/ here]. | ||
[[Image: | [[Image:Updated PfATP4 Activities.jpg|thumb|center|700px|Compounds so far Evaluated in the Kirk Ion Regulation Assay]] | ||
(Compounds that are inactive do not dissipate the plasma membrane Na<sup>+</sup> gradient or increase the plasma membrane pH gradient, consistent with them not inhibiting PfATP4 at the concentration tested. The other compounds dissipated the plasma membrane Na<sup>+</sup> gradient and increased the plasma membrane pH gradient at a concentration of 2 μM, consistent with them being PfATP4 inhibitors.) | |||
There is a correlation: compounds inactive in these assays are not potent vs the parasite. | |||
Testing was carried out vs. PfATP4-resistant mutants ([http://malaria.ourexperiment.org/biological_data/10943 ELN entry] and GHI [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/251 251]) in the laboratories of Kiaran Kirk and David Fidock [http://malaria.ourexperiment.org/uri/7a6 data here]. Also GHI[https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/251 251] | |||
[http://www.sciencedirect.com/science/article/pii/S2211320715300051 PfATP4 is the apparent target of a number of different chemotypes]. | |||
PfATP4 is implicated in the MoA of several other leading antimalarials in development: [http://www.sciencedirect.com/science/article/pii/S1931312813000358 the spiroindolone KAE609], the [http://www.nature.com/ncomms/2014/141125/ncomms6521/full/ncomms6521.html pyrazoleamides], the [http://www.pnas.org/content/111/50/E5455 dihydroisoquinolone (+)-SJ-733] and various [http://pubs.acs.org/doi/10.1021/cb500616x aminopyrazoles]. | |||
A striking diversity of other compounds ([http://onlinelibrary.wiley.com/doi/10.1111/mmi.12765/abstract from the malaria box]) behave in the same way ([http://malaria.ourexperiment.org/the_osm_blog/11547/The_Mysterious_World_of_ATP4_Inhibitors.html summarised here]), leading to the question: is PfATP4 really the target? | |||
There is no physical proof of binding between any of these compounds and PfATP4, because the protein has not yet been generated pure or crystallised. There is a [http://www.pnas.org/content/111/50/E5455 homology model]. | |||
Cross-resistance has been seen between parasites grown to be resistant to compound X (where there are mutations that are associated with PfATP4) and then tested with compound Y, including for the Series 4 compounds, as above. | |||
A current project strand is to develop a pharmacophore model that throws light on which compounds will be active in Kiaran's assay, and how they might be binding to PfATP4 (in part to allow us to de-prioritise the development of any more compounds having this same target). [http://malaria.ourexperiment.org/pharmacophore_modelling_/7971/Pharmacophore_Modelling_of_the_Malaria_Box_PfATP4_Active_Compounds.html An initial attempt at developing this model was unsuccessful] (i.e. not predictive - see the figure, where the "P Model predictions" correlate poorly with what was found in the ion regulation assay) possibly because the model did not allow for overlapping binding sites or take into consideration compound chirality. Model generation now needs to be re-attempted, and OSM needs community expertise in this area to proceed. | |||
( | |||
==hERG Activity== | ==hERG Activity== | ||
| Line 270: | Line 300: | ||
[[Image:Ubels Core Synthesis.png|thumb|center|650px|Summary of Synthetic Approach to Ether Series.]] | [[Image:Ubels Core Synthesis.png|thumb|center|650px|Summary of Synthetic Approach to Ether Series.]] | ||
[http://malaria.ourexperiment.org/triazolopyrazine_se/8217 The cyclisation was achieved], initially not in the ca. 50% yields reported by the original CRO. [http://malaria.ourexperiment.org/the_osm_blog/8894 Jo Ubels] has solved this, with input from others such as a student working with Patrick Thomson, [http://malaria.ourexperiment.org/triazolopyrazine_se/8335 Devon Scott]. | [http://malaria.ourexperiment.org/triazolopyrazine_se/8217 The cyclisation was achieved], initially not in the ca. 50% yields reported by the original CRO. [http://malaria.ourexperiment.org/the_osm_blog/8894 Jo Ubels] has solved this, with input from others such as a student working with Patrick Thomson, [http://malaria.ourexperiment.org/triazolopyrazine_se/8335 Devon Scott]. The boys at Sydney Grammar School have also [http://figshare.com/articles/Open_Source_Malaria_2015/1513845 studied the early stages of the synthesis]. | ||
The union of the alcohol fragment with the chloro-intermediate was for a long time not proceeding cleanly, and this [http://malaria.ourexperiment.org/data/files/9856/CRO%20Synth%20Series%204.doc was also found] by the CRO that first reported this route. Jo Ubels' [http://malaria.ourexperiment.org/the_osm_blog/9357 lit survey] found conditions involving a crown ether that has worked reliably. There have been periodic discussions about which alcohols to employ in the resulting synthetic scheme ([http://malaria.ourexperiment.org/the_osm_blog/9601Blog post from April 2014], GHI[https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/174 174]). | The union of the alcohol fragment with the chloro-intermediate was for a long time not proceeding cleanly, and this [http://malaria.ourexperiment.org/data/files/9856/CRO%20Synth%20Series%204.doc was also found] by the CRO that first reported this route. Jo Ubels' [http://malaria.ourexperiment.org/the_osm_blog/9357 lit survey] found conditions involving a crown ether that has worked reliably. There have been periodic discussions about which alcohols to employ in the resulting synthetic scheme ([http://malaria.ourexperiment.org/the_osm_blog/9601Blog post from April 2014], GHI[https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/174 174]). | ||
| Line 300: | Line 330: | ||
[[Image:Halogenated Core Synthesis.png|thumb|center|650px|Routes to the Halogenated Core Triazolopyrazine]] | [[Image:Halogenated Core Synthesis.png|thumb|center|650px|Routes to the Halogenated Core Triazolopyrazine]] | ||
Preliminary progress towards both has been made by Tom Macdonald (link to Honours thesis coming). The first steps (acid-mediated cyclizations) have been successfully carried out for Halogenation Routes [http://malaria.ourexperiment.org/triazolopyrazine_se/11112 1] and [http://malaria.ourexperiment.org/triazolopyrazine_se/11034 2]. The bromination step in Route 1 has reportedly been carried out (we lack the experimental write-up) and the halogenation in Route 2 was reportedly carried out in 58% yield ([http://malaria.ourexperiment.org/triazolopyrazine_se/11024 most recent lab book entry of presumably different attempt], [http://malaria.ourexperiment.org/triazolopyrazine_se/11287 more recent attempt by Alice]). | Preliminary progress towards both has been made by Tom Macdonald (link to Honours thesis coming). The first steps (acid-mediated cyclizations) have been successfully carried out for Halogenation Routes [http://malaria.ourexperiment.org/triazolopyrazine_se/11112 1] and [http://malaria.ourexperiment.org/triazolopyrazine_se/11034 2]. The bromination step in Route 1 has reportedly been carried out (we lack the experimental write-up) and the halogenation in Route 2 was reportedly carried out in 58% yield ([http://malaria.ourexperiment.org/triazolopyrazine_se/11024 most recent lab book entry of presumably different attempt], [http://malaria.ourexperiment.org/triazolopyrazine_se/11287 more recent attempt by Alice]). In 2016, [http://malaria.ourexperiment.org/triazolopyrazine_se/byuser/plus.google.com-109263878592735967907 Haochuan Mao completed a synthesis of the chloro-bromo compound], including securing its [http://malaria.ourexperiment.org/triazolopyrazine_se/14562/Crystal_Structure_Files_for_HM_32_the_bromochloroTP_core.html crystal structure], building on [http://malaria.ourexperiment.org/triazolopyrazine_se/byuser/plus.google.com-114378435836524935546 earlier work by Tianyi Zheng]. A [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/433 Haverford College superlab project also looked into this route]. | ||
===Fluoroalkene Isostere=== | ===Fluoroalkene Isostere=== | ||
| Line 338: | Line 368: | ||
==Desirable Compounds Not Yet Synthesised== | ==Desirable Compounds Not Yet Synthesised== | ||
[[Image:Tianyi compound.png|thumb|right|80px|Tianyi]] | |||
[[Image:Heterooxazole.png|thumb|right|80px|Heterooxazole]] | |||
[[Image:Heterooxadiazole.png|thumb|right|80px|Heterooxadiazole]] | |||
[[Image:HERG Evador.png|thumb|right|80px|hERG Evador]] | |||
[[Image:The Alkyl.png|thumb|right|80px|Alkyl]] | |||
[[Image:Internal Alkyne.png|thumb|right|80px|Internal Alkyne]] | |||
[[Image:Internal Alkene.png|thumb|right|80px|Internal Alkene]] | |||
[[Image:The 6.png|thumb|right|80px|The 6]] | |||
[[Image:The 8.png|thumb|right|80px|The 8]] | |||
[[Image:The Burns.png|thumb|right|80px|The Burns]] | |||
[[Image:The Nemesis.png|thumb|right|80px|The Nemesis]] | |||
[[Image:The Ketone.png|thumb|right|80px|The Ketone]] | |||
(Synthetic planning from Nov 2015 can be seen [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/358 here]) | |||
Side chain derived from phenylalanine: GHI [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/254 254], [http://malaria.ourexperiment.org/triazolopyrazine_se/10883/ ELN], [http://malaria.ourexperiment.org/osm_procedures/10911/OSMS261.html OSM-S-261]<br> | Side chain derived from phenylalanine: GHI [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/254 254], [http://malaria.ourexperiment.org/triazolopyrazine_se/10883/ ELN], [http://malaria.ourexperiment.org/osm_procedures/10911/OSMS261.html OSM-S-261]<br> | ||
ClC(C=C1)=CC=C1C2=NN=C3C=NC=C(OC[C@H](N)CC4=CC=CC=C4)N32 InChI=1S/C20H18ClN5O/c21-16-8-6-15(7-9-16)20-25-24-18-11-23-12-19(26(18)20)27-13-17(22)10-14-4-2-1-3-5-14/h1-9,11-12,17H,10,13,22H2/t17-/m1/s1 | ClC(C=C1)=CC=C1C2=NN=C3C=NC=C(OC[C@H](N)CC4=CC=CC=C4)N32 InChI=1S/C20H18ClN5O/c21-16-8-6-15(7-9-16)20-25-24-18-11-23-12-19(26(18)20)27-13-17(22)10-14-4-2-1-3-5-14/h1-9,11-12,17H,10,13,22H2/t17-/m1/s1 | ||
The '''Tianyi''' compound (contains methylated imidazopyrazine core, analogous to MMV669846): Discussion of Desirability: [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/358 GHI358], [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/390 GHI390] [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/410 Haverford Planning] | |||
OC(C1=CC=CC=C1)COC2=CN=CC3=NC(C)=C(C4=CC=C(OC(F)F)C=C4)N32 InChI=1S/C22H19F2N3O3/c1-14-21(16-7-9-17(10-8-16)30-22(23)24)27-19(26-14)11-25-12-20(27)29-13-18(28)15-5-3-2-4-6-15/h2-12,18,22,28H,13H2,1H3 VGNPLLBRBOUQAA-UHFFFAOYSA-N | |||
The '''Heterooxazole''' (unexplored amide isostere in side chain): [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/358 General discussion of the idea], [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/407 Proposed synthesis], [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/462 start of the synthesis], [http://malaria.ourexperiment.org/synthesis_of_hetero ELN] | |||
FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(C4=CN=C(C5=CC=CC=C5)O4)N32 (free acid) | |||
InChI=1S/C21H13F2N5O2/c22-21(23)29-15-8-6-13(7-9-15)19-27-26-18-12-24-10-16(28(18)19)17-11-25-20(30-17)14-4-2-1-3-5-14/h1-12,21H | |||
FBTBZFMQUBZDSN-UHFFFAOYSA-N | |||
The '''Heterooxadiazole''' (unexplored amide isostere in side chain): [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/358 General discussion of the idea], [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/453 Rationale], [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/406 Proposed synthesis], [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/446 Synthesis progress report by Haverford College], [http://malaria.ourexperiment.org/haverford_superlab_ Relevant ELN], [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/448 Revised synthetic route(s)], [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/447 Problems with imine formation], [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/449 Discussion of potentially anomalous NMR spectrum], [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/463 Second phase introduction] and [http://malaria.ourexperiment.org/hc_q2_het_oxadiazole new ELN] | |||
FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(C4=NC(C5=CC=CC=C5)=NO4)N32 | |||
InChI=1S/C20H12F2N6O2/c21-20(22)29-14-8-6-13(7-9-14)18-26-25-16-11-23-10-15(28(16)18)19-24-17(27-30-19)12-4-2-1-3-5-12/h1-11,20H | |||
HFPKPTYSFARKAA-UHFFFAOYSA-N | |||
The '''hERG Evador/Acid Test''' (carboxylic acid analogue of MMV688896): [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/390 Comment on desirability], [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/405 Synthesis proposal], [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/427 First synthesis attempt, summary and suggestions of ways forward] with [http://malaria.ourexperiment.org/chem_301_fall_2016_a ELN], [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/451 Discussion of synthetic roadblock], [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/452 Discussion of Rationale]. | |||
FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(OCC(C(O)=O)C4=CC=CC=C4)N32 | |||
InChI=1S/C21H16F2N4O4/c22-21(23)31-15-8-6-14(7-9-15)19-26-25-17-10-24-11-18(27(17)19)30-12-16(20(28)29)13-4-2-1-3-5-13/h1-11,16,21H,12H2,(H,28,29) | |||
FICNFVPYGUTYFV-UHFFFAOYSA-N | |||
The '''Alkyl''' (Test to see if heteroatoms are needed on left-hand-side). Length may need tweaking - MMV669304 is potent): [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/403 1st proposed synthesis] with [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/424 commencement] and [http://malaria.ourexperiment.org/morrison_and_milder ELN], [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/418 2nd proposed synthesis] with [http://malaria.ourexperiment.org/synthesis_of_the_al ELN], | |||
FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(CCC4=CC=CC=C4)N32 | |||
InChI=1S/C20H16F2N4O/c21-20(22)27-17-10-7-15(8-11-17)19-25-24-18-13-23-12-16(26(18)19)9-6-14-4-2-1-3-5-14/h1-5,7-8,10-13,20H,6,9H2 | |||
QKTHSIJWBSHMDZ-UHFFFAOYSA-N | |||
The '''Internal Alkyne''' (related to alkyl): [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/423 Opening synthesis], [http://malaria.ourexperiment.org/internal_alkyl ELN], [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/455 discussion of rationale] | |||
FC(F)OC1=CC=C(C2=NN=C3C=NC=C(C#CC4=CC=CC=C4)N32)C=C1 | |||
InChI=1S/C20H12F2N4O/c21-20(22)27-17-10-7-15(8-11-17)19-25-24-18-13-23-12-16(26(18)19)9-6-14-4-2-1-3-5-14/h1-5,7-8,10-13,20H | |||
KNVACGGMAGPRRW-UHFFFAOYSA-N | |||
The '''Internal Alkene/Reduced Alkyl''' (synthetically related to alkyne and alkyl compounds; [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/457 discussion of rationale of all three]; The SMILES string is only for the ''cis'' isomer: [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/403 Discussed], [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/431 starting synthesis], [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/454 Discussion of NMR spectrum of intermediate], [http://malaria.ourexperiment.org/reduced_alkyl ELN] | |||
FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(/C=C\C4=CC=CC=C4)N32 | |||
InChI=1S/C20H14F2N4O/c21-20(22)27-17-10-7-15(8-11-17)19-25-24-18-13-23-12-16(26(18)19)9-6-14-4-2-1-3-5-14/h1-13,20H/b9-6- | |||
SGPCUHISMIPDDU-TWGQIWQCSA-N | |||
The '''6''' (probe of tolerance to ring substitution): [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/401 Synthesis proposal], [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/428 Start of synthesis], [http://malaria.ourexperiment.org/synthesis_of__the_6 ELN]. [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/461 Second phase Haverford attempt] with [http://malaria.ourexperiment.org/haverford_college_s second ELN] | |||
FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC(C)=C(OCC(O)C4=CC=CC=C4)N32 | |||
InChI=1S/C21H18F2N4O3/c1-13-20(29-12-17(28)14-5-3-2-4-6-14)27-18(11-24-13)25-26-19(27)15-7-9-16(10-8-15)30-21(22)23/h2-11,17,21,28H,12H2,1H3 | |||
ZYFKQWHTACKGJR-UHFFFAOYSA-N | |||
The '''8''' (also a probe of tolerance to ring substitution): [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/402 Haverford synthesis plan], [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/429 Start of Haverford synthesis], [http://malaria.ourexperiment.org/superlabthe8 ELN], [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/444 Rationale], [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/456 Summary and closure of first attempt], [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/461 cross-link to new attempt at 6- and 8-] | |||
FC(F)OC(C=C1)=CC=C1C2=NN=C3C(C)=NC=C(OCC(O)C4=CC=CC=C4)N32 | |||
InChI=1S/C21H18F2N4O3/c1-13-19-25-26-20(15-7-9-16(10-8-15)30-21(22)23)27(19)18(11-24-13)29-12-17(28)14-5-3-2-4-6-14/h2-11,17,21,28H,12H2,1H3 | |||
ASWYMQOLUDKKDJ-UHFFFAOYSA-N | |||
The '''Burns''' (amide isostere): [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/404 Proposed synthesis from Haverford College], [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/422 Start of synthesis], Discussion of results of intermediate steps [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/438 here] and [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/450 here], [http://malaria.ourexperiment.org/the_burns ELN] including [http://malaria.ourexperiment.org/the_burns/14906/Synthesis_of__Nbenzyl16chloropyrazin2yl222trifluoroethan1amine_EVKCN_14.html summary of outcomes of first attempt]. [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/459 Start of second attempt] with new [http://malaria.ourexperiment.org/hc_superlab_ ELN]. | |||
FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(C(C(F)(F)F)NCC4=CC=CC=C4)N32 | |||
InChI=1S/C21H16F5N5O/c22-20(23)32-15-8-6-14(7-9-15)19-30-29-17-12-27-11-16(31(17)19)18(21(24,25)26)28-10-13-4-2-1-3-5-13/h1-9,11-12,18,20,28H,10H2 | |||
NYRGMZUFRGWPBF-UHFFFAOYSA-N | |||
The '''Nemesis''' (deplanarisation of right-hand-side): [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/409 Haverford College synthetic plan], [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/430 Start of first Haverford synthesis], [http://malaria.ourexperiment.org/hcnemesis16 ELN], [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/466 Second phase of synthesis with same ELN]. | |||
OC(C1=CC=CC=C1)COC2=CN=CC3=NN=C(C4CNCCC4)N32 | |||
InChI=1S/C18H21N5O2/c24-15(13-5-2-1-3-6-13)12-25-17-11-20-10-16-21-22-18(23(16)17)14-7-4-8-19-9-14/h1-3,5-6,10-11,14-15,19,24H,4,7-9,12H2 | |||
SNDLBGDEOQPBOQ-UHFFFAOYSA-N | |||
The '''Ketone''' (analog of amide linker): [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/408 Haverford College proposed syntheses], [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/425 Start of synthesis], [http://malaria.ourexperiment.org/ketone_synthesis ELN]. Also "ketone linker" folder in [https://au-mynotebook.labarchives.com/share/Edwin%2520Tse/MTYzLjh8MTUwOS8xMjYvVHJlZU5vZGUvMjUxNDEwNDEwOHw0MTUuOA== Ed Tse's lab notebook]. [https://github.com/OpenSourceMalaria/OSM_To_Do_List/issues/460 Second Haverford College campaign] with [http://malaria.ourexperiment.org/ketone_q2 new ELN]. | |||
FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(C(CC4=CC=CC=C4)=O)N32 | |||
InChI=1S/C20H14F2N4O2/c21-20(22)28-15-8-6-14(7-9-15)19-25-24-18-12-23-11-16(26(18)19)17(27)10-13-4-2-1-3-5-13/h1-9,11-12,20H,10H2 | |||
KALFSUSYOZTBSP-UHFFFAOYSA-N | |||
==Strings for Google== | ==Strings for Google== | ||
Use this section to paste strings to make the page more discoverable. | Use this section to paste strings to make the page more discoverable.<br> | ||
Format (to allow creation of SDF): MMV Code (OSM Code and link to page) SMILES InChI InChIKey Potency in μmol | Format (to allow creation of SDF): MMV Code (OSM Code and link to page) SMILES InChI InChIKey Potency in μmol | ||
| Line 361: | Line 464: | ||
'''MMV668962''' FC(F)OC(C=C1)=CC=C1C2=NN=C3N2CCN(C(CN)=O)C3 InChI=1S/C14H15F2N5O2/c15-14(16)23-10-3-1-9(2-4-10)13-19-18-11-8-20(12(22)7-17)5-6-21(11)13/h1-4,14H,5-8,17H2 <br> | '''MMV668962''' FC(F)OC(C=C1)=CC=C1C2=NN=C3N2CCN(C(CN)=O)C3 InChI=1S/C14H15F2N5O2/c15-14(16)23-10-3-1-9(2-4-10)13-19-18-11-8-20(12(22)7-17)5-6-21(11)13/h1-4,14H,5-8,17H2 <br> | ||
'''MMV669000''' ([http://malaria.ourexperiment.org/osm_procedures/9606/Preparation_of_OSMS177.html OSM-S-177]) O=C(N1CC(C=CC=C2)=C2C1)C3=CN=CC4=NN=C(C5=CC=C(OC(F)F)C=C5)N43 InChI=1S/C21H15F2N5O2/c22-21(23)30-16-7-5-13(6-8-16)19-26-25-18-10-24-9-17(28(18)19)20(29)27-11-14-3-1-2-4-15(14)12-27/h1-10,21H,11-12H2 <br> | '''MMV669000''' ([http://malaria.ourexperiment.org/osm_procedures/9606/Preparation_of_OSMS177.html OSM-S-177]) O=C(N1CC(C=CC=C2)=C2C1)C3=CN=CC4=NN=C(C5=CC=C(OC(F)F)C=C5)N43 InChI=1S/C21H15F2N5O2/c22-21(23)30-16-7-5-13(6-8-16)19-26-25-18-10-24-9-17(28(18)19)20(29)27-11-14-3-1-2-4-15(14)12-27/h1-10,21H,11-12H2 <br> | ||
'''MMV669001''' ([http://malaria.ourexperiment.org/osm_procedures/9608/Preparation_of_OSMS178.html OSM-S-178]) | '''MMV669001''' ([http://malaria.ourexperiment.org/osm_procedures/9608/Preparation_of_OSMS178.html OSM-S-178]) c1ncc2n(c1C(NCc1ccc(cc1)Cl)=O)c(nn2)c1ccc(cc1)OC(F)F InChI=1S/C20H14ClF2N5O2/c21-14-5-1-12(2-6-14)9-25-19(29)16-10-24-11-17-26-27-18(28(16)17)13-3-7-15(8-4-13)30-20(22)23/h1-8,10-11,20H,9H2,(H,25,29) InChI=1S/C20H14ClF2N5O2/c21-14-5-1-12(2-6-14)9-25-19(29)16-10-24-11-17-26-27-18(28(16)17)13-3-7-15(8-4-13)30-20(22)23/h1-8,10-11,20H,9H2,(H,25,29) <br> | ||
'''MMV669002''' in amide scheme <br> | '''MMV669002''' in amide scheme <br> | ||
'''MMV669003''' ([http://malaria.ourexperiment.org/osm_procedures/9609/Preparation_of_OSMS179.html OSM-S-179]) in amide scheme <br> | '''MMV669003''' ([http://malaria.ourexperiment.org/osm_procedures/9609/Preparation_of_OSMS179.html OSM-S-179]) in amide scheme <br> | ||
'''MMV669006''' FC1=C(F)C=CC(CCOC2=CN=CC3=NN=C(NC4=CC=C(F)C(F)=C4)N32)=C1 InChI=1S/C19H13F4N5O/c20-13-3-1-11(7-15(13)22)5-6-29-18-10-24-9-17-26-27-19(28(17)18)25-12-2-4-14(21)16(23)8-12/h1-4,7-10H,5-6H2,(H,25,27) <br> | '''MMV669006''' FC1=C(F)C=CC(CCOC2=CN=CC3=NN=C(NC4=CC=C(F)C(F)=C4)N32)=C1 InChI=1S/C19H13F4N5O/c20-13-3-1-11(7-15(13)22)5-6-29-18-10-24-9-17-26-27-19(28(17)18)25-12-2-4-14(21)16(23)8-12/h1-4,7-10H,5-6H2,(H,25,27) <br> | ||
'''MMV669008''' FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(N4CC(OC5=CC(F)=C(F)C=C5)C4)N32 InChI=1S/C21H15F4N5O2/c22-16-6-5-14(7-17(16)23)31-15-10-29(11-15)19-9-26-8-18-27-28-20(30(18)19)12-1-3-13(4-2-12)32-21(24)25/h1-9,15,21H,10-11H2 <br> | '''MMV669008''' FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(N4CC(OC5=CC(F)=C(F)C=C5)C4)N32 InChI=1S/C21H15F4N5O2/c22-16-6-5-14(7-17(16)23)31-15-10-29(11-15)19-9-26-8-18-27-28-20(30(18)19)12-1-3-13(4-2-12)32-21(24)25/h1-9,15,21H,10-11H2 <br> | ||
'''MMV669009''' FC1=C(F)C=CC(CCOC2=CN=CC3=NN=C(N4CCCCC4)N32)=C1 InChI=1S/C18H19F2N5O/c19-14-5-4-13(10-15(14)20)6-9-26-17-12-21-11-16-22-23-18(25(16)17)24-7-2-1-3-8-24/h4-5,10-12H,1-3,6-9H2 <br> | '''MMV669009''' FC1=C(F)C=CC(CCOC2=CN=CC3=NN=C(N4CCCCC4)N32)=C1 InChI=1S/C18H19F2N5O/c19-14-5-4-13(10-15(14)20)6-9-26-17-12-21-11-16-22-23-18(25(16)17)24-7-2-1-3-8-24/h4-5,10-12H,1-3,6-9H2 <br> | ||
'''MMV669010''' ([http://malaria.ourexperiment.org/osm_procedures/9610/Preparation_of_OSMS180.html OSM-S-180]) | '''MMV669010''' ([http://malaria.ourexperiment.org/osm_procedures/9610/Preparation_of_OSMS180.html OSM-S-180]) c1ncc2n(c1C(N1CCN(CC1)c1ncccc1)=O)c(nn2)c1ccc(cc1)OC(F)F InChI=1S/C23H22F2N8O2/c24-23(25)35-17-6-4-16(5-7-17)21-30-29-20-14-26-13-18(33(20)21)22(34)28-15-31-9-11-32(12-10-31)19-3-1-2-8-27-19/h1-8,13-14,23H,9-12,15H2,(H,28,34) UYWFTHNNGLIMAY-UHFFFAOYSA-N <br> | ||
'''MMV669011''' ([http://malaria.ourexperiment.org/osm_procedures/9611/Preparation_of_OSMS181.html OSM-S-181]) | '''MMV669011''' ([http://malaria.ourexperiment.org/osm_procedures/9611/Preparation_of_OSMS181.html OSM-S-181]) c1ncc2n(c1C(NCCN1CCOCC1)=O)c(nn2)c1ccc(cc1)OC(F)F InChI=1S/C19H20F2N6O3/c20-19(21)30-14-3-1-13(2-4-14)17-25-24-16-12-22-11-15(27(16)17)18(28)23-5-6-26-7-9-29-10-8-26/h1-4,11-12,19H,5-10H2,(H,23,28) LYBWZYLEKBKOOG-UHFFFAOYSA-N <br> | ||
'''MMV669020''' ([http://malaria.ourexperiment.org/osm_procedures/9624/Preparation_of_OSMS185.html OSM-S-185]) | '''MMV669020''' ([http://malaria.ourexperiment.org/osm_procedures/9624/Preparation_of_OSMS185.html OSM-S-185]) c1ncc2n(c1C(N1CCOCC1)=O)c(nn2)c1ccc(cc1)OC(F)F InChI=1S/C17H15F2N5O3/c18-17(19)27-12-3-1-11(2-4-12)15-22-21-14-10-20-9-13(24(14)15)16(25)23-5-7-26-8-6-23/h1-4,9-10,17H,5-8H2 LFRCBCKOIXZMLV-UHFFFAOYSA-N <br> | ||
'''MMV669021''' in amide scheme <br> | '''MMV669021''' c1ncc2n(c1C(N1CCCC1)=O)c(nn2)c1ccc(cc1)OC(F)F in amide scheme <br> | ||
'''MMV669022''' ([http://malaria.ourexperiment.org/osm_procedures/9621/Preparation_of_OSMS182.html OSM-S-182]) | '''MMV669022''' ([http://malaria.ourexperiment.org/osm_procedures/9621/Preparation_of_OSMS182.html OSM-S-182]) InChI=1S/C17H15F2N5O2/c18-17(19)26-12-5-3-11(4-6-12)15-22-21-14-10-20-9-13(24(14)15)16(25)23-7-1-2-8-23/h3-6,9-10,17H,1-2,7-8H2 HDIHMYGJUUDJKO-UHFFFAOYSA-N <br> | ||
'''MMV669023''' ([http://malaria.ourexperiment.org/osm_procedures/9623/Preparation_of_OSMS183.html OSM-S-184]) in amide scheme <br> | '''MMV669023''' ([http://malaria.ourexperiment.org/osm_procedures/9623/Preparation_of_OSMS183.html OSM-S-184]) in amide scheme <br> | ||
'''MMV669024''' ([http://malaria.ourexperiment.org/osm_procedures/9625/Preparation_of_OSMS186.html OSM-S-186]) in amide scheme <br> | '''MMV669024''' ([http://malaria.ourexperiment.org/osm_procedures/9625/Preparation_of_OSMS186.html OSM-S-186]) in amide scheme <br> | ||
| Line 396: | Line 499: | ||
'''MMV669844''' ([http://malaria.ourexperiment.org/osm_procedures/9951/OSMS218.html OSM-S-218]) [C@H](COc1cncc2n1c(nn2)c1ccc(cc1)C#N)(c1cc(c(cc1)F)F)OC | '''MMV669844''' ([http://malaria.ourexperiment.org/osm_procedures/9951/OSMS218.html OSM-S-218]) [C@H](COc1cncc2n1c(nn2)c1ccc(cc1)C#N)(c1cc(c(cc1)F)F)OC | ||
'''MMV669846''' FC1=C(F)C=CC(CCOC2=CN=CC3=NC=C(C4=CC=C(Cl)C=C4)N32)=C1 InChI=1S/C20H14ClF2N3O/c21-15-4-2-14(3-5-15)18-10-25-19-11-24-12-20(26(18)19)27-8-7-13-1-6-16(22)17(23)9-13/h1-6,9-12H,7-8H2 <br> | '''MMV669846''' ([http://malaria.ourexperiment.org/osm_procedures/11554/Preparation_of_OSMS273.html OSM-S-273]) FC1=C(F)C=CC(CCOC2=CN=CC3=NC=C(C4=CC=C(Cl)C=C4)N32)=C1 InChI=1S/C20H14ClF2N3O/c21-15-4-2-14(3-5-15)18-10-25-19-11-24-12-20(26(18)19)27-8-7-13-1-6-16(22)17(23)9-13/h1-6,9-12H,7-8H2 MQHQNFQVEXYAMO-UHFFFAOYSA-N 0.110 <br> | ||
'''MMV669848''' FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(CN4CC(C=CC=C5)=C5C4)N32 InChI=1S/C21H17F2N5O/c22-21(23)29-18-7-5-14(6-8-18)20-26-25-19-10-24-9-17(28(19)20)13-27-11-15-3-1-2-4-16(15)12-27/h1-10,21H,11-13H2 <br> | '''MMV669848''' FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(CN4CC(C=CC=C5)=C5C4)N32 InChI=1S/C21H17F2N5O/c22-21(23)29-18-7-5-14(6-8-18)20-26-25-19-10-24-9-17(28(19)20)13-27-11-15-3-1-2-4-16(15)12-27/h1-10,21H,11-13H2 <br> | ||
'''MMV669849''' in amide scheme <br> | '''MMV669849''' in amide scheme <br> | ||
| Line 403: | Line 506: | ||
'''MMV670246''' FC(F)OC(C=C1)=CC=C1C2=NN=C3N2C(C(NC4=CC=C(Cl)C=C4)=O)=CN=C3 InChI=1S/C19H12ClF2N5O2/c20-12-3-5-13(6-4-12)24-18(28)15-9-23-10-16-25-26-17(27(15)16)11-1-7-14(8-2-11)29-19(21)22/h1-10,19H,(H,24,28) <br> | '''MMV670246''' FC(F)OC(C=C1)=CC=C1C2=NN=C3N2C(C(NC4=CC=C(Cl)C=C4)=O)=CN=C3 InChI=1S/C19H12ClF2N5O2/c20-12-3-5-13(6-4-12)24-18(28)15-9-23-10-16-25-26-17(27(15)16)11-1-7-14(8-2-11)29-19(21)22/h1-10,19H,(H,24,28) <br> | ||
'''MMV670249''' FC1=C(F)C=CC(CCOC2=CN=CC3=NN=C(N4CCC5=C4C=CC(F)=C5)N32)=C1 InChI=1S/C21H16F3N5O/c22-15-2-4-18-14(10-15)5-7-28(18)21-27-26-19-11-25-12-20(29(19)21)30-8-6-13-1-3-16(23)17(24)9-13/h1-4,9-12H,5-8H2 <br> | '''MMV670249''' FC1=C(F)C=CC(CCOC2=CN=CC3=NN=C(N4CCC5=C4C=CC(F)=C5)N32)=C1 InChI=1S/C21H16F3N5O/c22-15-2-4-18-14(10-15)5-7-28(18)21-27-26-19-11-25-12-20(29(19)21)30-8-6-13-1-3-16(23)17(24)9-13/h1-4,9-12H,5-8H2 <br> | ||
'''MMV670250''' FC1=C(F)C=CC(CCOC2=CN=CC3=CN=C(C4=CC=C(Cl)C=C4)N32)=C1 InChI=1S/C20H14ClF2N3O/c21-15-4-2-14(3-5-15)20-25-11-16-10-24-12-19(26(16)20)27-8-7-13-1-6-17(22)18(23)9-13/h1-6,9-12H,7-8H2 <br> | '''MMV670250''' ([http://malaria.ourexperiment.org/osm_procedures/11558/Preparation_of_OSMS274.html OSM-S-274]) FC1=C(F)C=CC(CCOC2=CN=CC3=CN=C(C4=CC=C(Cl)C=C4)N32)=C1 InChI=1S/C20H14ClF2N3O/c21-15-4-2-14(3-5-15)20-25-11-16-10-24-12-19(26(16)20)27-8-7-13-1-6-17(22)18(23)9-13/h1-6,9-12H,7-8H2 JHORUMGXLQTHAD-UHFFFAOYSA-N 0.830 <br> | ||
'''MMV670437''' FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(OC[C@H](N(C)C)C4=CC(F)=C(F)C=C4)N32 InChI=1S/C22H19F4N5O2/c1-30(2)18(14-5-8-16(23)17(24)9-14)12-32-20-11-27-10-19-28-29-21(31(19)20)13-3-6-15(7-4-13)33-22(25)26/h3-11,18,22H,12H2,1-2H3/t18-/m0/s1 <br> | '''MMV670437''' FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(OC[C@H](N(C)C)C4=CC(F)=C(F)C=C4)N32 InChI=1S/C22H19F4N5O2/c1-30(2)18(14-5-8-16(23)17(24)9-14)12-32-20-11-27-10-19-28-29-21(31(19)20)13-3-6-15(7-4-13)33-22(25)26/h3-11,18,22H,12H2,1-2H3/t18-/m0/s1 <br> | ||
'''MMV670438''' FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OCC4(COC4)C5=CC=C(F)C(F)=C5 InChI=1S/C22H16F4N4O3/c23-16-6-3-14(7-17(16)24)22(10-31-11-22)12-32-19-9-27-8-18-28-29-20(30(18)19)13-1-4-15(5-2-13)33-21(25)26/h1-9,21H,10-12H2 <br> | '''MMV670438''' FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OCC4(COC4)C5=CC=C(F)C(F)=C5 InChI=1S/C22H16F4N4O3/c23-16-6-3-14(7-17(16)24)22(10-31-11-22)12-32-19-9-27-8-18-28-29-20(30(18)19)13-1-4-15(5-2-13)33-21(25)26/h1-9,21H,10-12H2 <br> | ||
| Line 455: | Line 558: | ||
'''MMV672688''' FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OCC4=CC=CC5=C4C=CC=C5 InChI=1S/C23H16F2N4O2/c24-23(25)31-18-10-8-16(9-11-18)22-28-27-20-12-26-13-21(29(20)22)30-14-17-6-3-5-15-4-1-2-7-19(15)17/h1-13,23H,14H2 <br> | '''MMV672688''' FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OCC4=CC=CC5=C4C=CC=C5 InChI=1S/C23H16F2N4O2/c24-23(25)31-18-10-8-16(9-11-18)22-28-27-20-12-26-13-21(29(20)22)30-14-17-6-3-5-15-4-1-2-7-19(15)17/h1-13,23H,14H2 <br> | ||
'''MMV672689''' FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(O[C@H]4CN(C5CC5)[C@@H]4C6=CC(F)=C(F)C=C6)N32 InChI=1S/C24H19F4N5O2/c25-17-8-3-14(9-18(17)26)22-19(12-32(22)15-4-5-15)35-21-11-29-10-20-30-31-23(33(20)21)13-1-6-16(7-2-13)34-24(27)28/h1-3,6-11,15,19,22,24H,4-5,12H2/t19-,22+/m0/s1 <br> | '''MMV672689''' FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(O[C@H]4CN(C5CC5)[C@@H]4C6=CC(F)=C(F)C=C6)N32 InChI=1S/C24H19F4N5O2/c25-17-8-3-14(9-18(17)26)22-19(12-32(22)15-4-5-15)35-21-11-29-10-20-30-31-23(33(20)21)13-1-6-16(7-2-13)34-24(27)28/h1-3,6-11,15,19,22,24H,4-5,12H2/t19-,22+/m0/s1 <br> | ||
'''MMV672723''' FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OCC(O)(C)C4=CC=C(F)C(F)=C4 InChI=1S/C21H16F4N4O3/c1-21(30,13-4-7-15(22)16(23)8-13)11-31-18-10-26-9-17-27-28-19(29(17)18)12-2-5-14(6-3-12)32-20(24)25/h2-10,20,30H,11H2,1H3 <br> | '''MMV672723''' FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OCC(O)(C)C4=CC=C(F)C(F)=C4 InChI=1S/C21H16F4N4O3/c1-21(30,13-4-7-15(22)16(23)8-13)11-31-18-10-26-9-17-27-28-19(29(17)18)12-2-5-14(6-3-12)32-20(24)25/h2-10,20,30H,11H2,1H3 <br> | ||
'''MMV672725''' (mentioned in Jo Ubels Ether scheme) <br> | '''MMV672725''' (mentioned in Jo Ubels Ether scheme) <br> | ||
| Line 470: | Line 572: | ||
'''MMV672990''' FC(F)OC(C=C1)=CC=C1C2=NN=C3N2C=C(NC(CC4=CC=CC(Cl)=C4)=O)N=C3 InChI=1S/C20H14ClF2N5O2/c21-14-3-1-2-12(8-14)9-18(29)25-16-11-28-17(10-24-16)26-27-19(28)13-4-6-15(7-5-13)30-20(22)23/h1-8,10-11,20H,9H2,(H,25,29) <br> | '''MMV672990''' FC(F)OC(C=C1)=CC=C1C2=NN=C3N2C=C(NC(CC4=CC=CC(Cl)=C4)=O)N=C3 InChI=1S/C20H14ClF2N5O2/c21-14-3-1-2-12(8-14)9-18(29)25-16-11-28-17(10-24-16)26-27-19(28)13-4-6-15(7-5-13)30-20(22)23/h1-8,10-11,20H,9H2,(H,25,29) <br> | ||
'''MMV672992''' N#CC(C=C1)=CC=C1C2=NN=C3C=CC(NCCC4=CN=CC=C4)=NN32 InChI=1S/C19H15N7/c20-12-14-3-5-16(6-4-14)19-24-23-18-8-7-17(25-26(18)19)22-11-9-15-2-1-10-21-13-15/h1-8,10,13H,9,11H2,(H,22,25) <br> | '''MMV672992''' N#CC(C=C1)=CC=C1C2=NN=C3C=CC(NCCC4=CN=CC=C4)=NN32 InChI=1S/C19H15N7/c20-12-14-3-5-16(6-4-14)19-24-23-18-8-7-17(25-26(18)19)22-11-9-15-2-1-10-21-13-15/h1-8,10,13H,9,11H2,(H,22,25) <br> | ||
'''JS 11-1''' ClC1=CC=CC2=NN=C(C3=CC=C(C#N)C=C3)N21 InChI=1S/C13H7ClN4/c14-11-2-1-3-12-16-17-13(18(11)12)10-6-4-9(8-15)5-7-10/h1-7H ROKQDRVNUIOPKS-UHFFFAOYSA-N >50 <br> | |||
'''JS 19-1''' ClC(C=CC=C1)=C1CCOC2=CC=CC3=NN=C(C4=CC=C(C#N)C=C4)N32 InChI=1S/C21H15ClN4O/c22-18-5-2-1-4-16(18)12-13-27-20-7-3-6-19-24-25-21(26(19)20)17-10-8-15(14-23)9-11-17/h1-11H,12-13H2 APTFJFHOJWNXBX-UHFFFAOYSA-N 5.2 <br> | |||
'''JS 20-1''' N#CC(C=C1)=CC=C1C2=NN=C3C=CC=C(OCCC4=CC=CC=C4)N32 InChI=1S/C21H16N4O/c22-15-17-9-11-18(12-10-17)21-24-23-19-7-4-8-20(25(19)21)26-14-13-16-5-2-1-3-6-16/h1-12H,13-14H2 PLXIJFIDAXQTSX-UHFFFAOYSA-N 6.8 <br> | |||
'''JS 21-1''' FC(C=C1)=C(F)C=C1CCOC2=CC=CC3=NN=C(C4=CC=C(C#N)C=C4)N32 InChI=1S/C21H14F2N4O/c22-17-9-6-14(12-18(17)23)10-11-28-20-3-1-2-19-25-26-21(27(19)20)16-7-4-15(13-24)5-8-16/h1-9,12H,10-11H2 JCMCKOQZSFBPIN-UHFFFAOYSA-N 3.5 <br> | |||
'''JS 16-1''' O=C(NC1=CC=NC(C(F)(F)F)=C1)C2=CC=CC3=NN=C(C4=CC=C(C#N)C=C4)N32 InChI=1S/C20H11F3N6O/c21-20(22,23)16-10-14(8-9-25-16)26-19(30)15-2-1-3-17-27-28-18(29(15)17)13-6-4-12(11-24)5-7-13/h1-10H,(H,25,26,30) PGPJMFWOQCDCCV-UHFFFAOYSA-N >50 <br> | |||
'''MMV688896''' OC(C1=CC=CC=C1)COC2=CN=CC3=NN=C(C4=CC=C(OC(F)F)C=C4)N32 InChI=1S/C20H16F2N4O3/c21-20(22)29-15-8-6-14(7-9-15)19-25-24-17-10-23-11-18(26(17)19)28-12-16(27)13-4-2-1-3-5-13/h1-11,16,20,27H,12H2 GEHCMLKWLSBHDX-UHFFFAOYSA-N 0.27 <br> | |||
'''MMV688894''' OC(C1=CC=CC=C1)COC2=CN=CC3=NN=C(C4=CC=CC=C4)N32 InChI=1S/C19H16N4O2/c24-16(14-7-3-1-4-8-14)13-25-18-12-20-11-17-21-22-19(23(17)18)15-9-5-2-6-10-15/h1-12,16,24H,13H2 HQWUAOFYEUBLBZ-UHFFFAOYSA-N >5 <br> | |||
'''MMV688895''' OC(C1=CC=CC=C1)COC2=CN=CC3=NN=C(C4=CN=C(C(F)(F)F)C=C4)N32 InChI=1S/C19H14F3N5O2/c20-19(21,22)15-7-6-13(8-24-15)18-26-25-16-9-23-10-17(27(16)18)29-11-14(28)12-4-2-1-3-5-12/h1-10,14,28H,11H2 YUTTVQUFLHMVCQ-UHFFFAOYSA-N 3.462, 3.665 <br> | |||
'''MMV688892''' OC(C1=CC=CC=C1)COC2=CN=CC3=NN=C(C4CCCCC4)N32 InChI=1S/C19H22N4O2/c24-16(14-7-3-1-4-8-14)13-25-18-12-20-11-17-21-22-19(23(17)18)15-9-5-2-6-10-15/h1,3-4,7-8,11-12,15-16,24H,2,5-6,9-10,13H2 LWDWZIVAQMSZHL-UHFFFAOYSA-N >5 <br> | |||
'''MMV688893''' CCC1=NN=C2N1C(OCC(O)C3=CC=CC=C3)=CN=C2 InChI=1S/C15H16N4O2/c1-2-13-17-18-14-8-16-9-15(19(13)14)21-10-12(20)11-6-4-3-5-7-11/h3-9,12,20H,2,10H2,1H3 ZMELTVBBQIUHNR-UHFFFAOYSA-N >10 <br> | |||
'''MMV675718''' ([http://malaria.ourexperiment.org/osm_procedures/9868/Preparation_of_OSMS201.html OSM-S-201]) <br> | '''MMV675718''' ([http://malaria.ourexperiment.org/osm_procedures/9868/Preparation_of_OSMS201.html OSM-S-201]) <br> | ||
| Line 480: | Line 593: | ||
'''MMV675951''' ([http://malaria.ourexperiment.org/osm_procedures/10869/Preparation_of_OSMS256.html OSM-S-256]) <br> | '''MMV675951''' ([http://malaria.ourexperiment.org/osm_procedures/10869/Preparation_of_OSMS256.html OSM-S-256]) <br> | ||
'''MMV675952''' ([http://malaria.ourexperiment.org/osm_procedures/10870/Preparation_of_OSMS257.html OSM-S-257]) <br> | '''MMV675952''' ([http://malaria.ourexperiment.org/osm_procedures/10870/Preparation_of_OSMS257.html OSM-S-257]) <br> | ||
'''MMV675959''' ([http://malaria.ourexperiment.org/osm_procedures/10908/OSMS259.html OSM-S-259]) <br> | |||
'''MMV675959''' ([http://malaria.ourexperiment.org/osm_procedures/10908/OSMS259.html OSM-S-259]) FC1=CC(COC2=CN=CC3=NN=C(C4=CC=C(C#N)C=C4)N32)=CC=C1F InChI=1S/C19H11F2N5O/c20-15-6-3-13(7-16(15)21)11-27-18-10-23-9-17-24-25-19(26(17)18)14-4-1-12(8-22)2-5-14/h1-7,9-10H,11H2 AUBWBFLGMHEXIB-UHFFFAOYSA-N 0.452 <br> | |||
'''MMV675960''' ([http://malaria.ourexperiment.org/osm_procedures/10910/OSMS260.html OSM-S-260]) <br> | '''MMV675960''' ([http://malaria.ourexperiment.org/osm_procedures/10910/OSMS260.html OSM-S-260]) <br> | ||
'''MMV675961''' ([http://malaria.ourexperiment.org/osm_procedures/10921/OSMS262.html OSM-S-262]) <br> | '''MMV675961''' ([http://malaria.ourexperiment.org/osm_procedures/10921/OSMS262.html OSM-S-262]) <br> | ||
| Line 496: | Line 611: | ||
([http://malaria.ourexperiment.org/osm_procedures/9963/Preparation_of_OSMS219.html OSM-S-219]) <br> | ([http://malaria.ourexperiment.org/osm_procedures/9963/Preparation_of_OSMS219.html OSM-S-219]) <br> | ||
([http://malaria.ourexperiment.org/osm_procedures/9966/Preparation_of_OSMS220.html OSM-S-220]) <br> | ([http://malaria.ourexperiment.org/osm_procedures/9966/Preparation_of_OSMS220.html OSM-S-220]) <br> | ||
<!-- END OF FILE --> | |||
Latest revision as of 19:36, 8 August 2017
Important Note - this page is no longer being updated. The active wiki is here.
Open Source Malaria Series 4: The Triazolopyrazine (TP) Series
Resources
Molecules and How to Visualize Them
OSIRIS DataWarrior (general purpose open-source chemoinformatics software)
Google Sheet of all OSM Compounds (Can filter Series 4).
Sister sheet that explains column headings (i.e. assays used).
Visualisation of OSM molecules on Cheminfo. The data may be visualized with Vortex. The Series 4 SDF can be generated on Cheminfo - here is a downloadable version from May 2016.
Lab Notebooks
The principal Labtrove ELN
Ed Tse's Labarchives ELN
Jasper Tyler's Lab Notebook
Chase Smith's Labarchives ELN
Old document from CRO describing synthetic chemistry of some members of series
Papers
First paper based on this series is being written up here. Relevant 2016 review from MMV. 2017 New developments in anti-malarial target candidate and product profiles
How To Join In
The To Do List
How to involve yourself in OSM is described in the Landing Page under "Join the Team".
Document Archive
Initial briefing document on the series written for MMV and a PDF summary of pharmacokinetics and efficacy; largely folded into the page below. (A minor error in the briefing document referring to the amides has been clarified.)
PDF of the Compounds Originally Inherited from MMV including structures and potencies ([http://malaria.ourexperiment.org/osdd_malaria_shared/8106/MMV_triazolopyrazine_data.html older version of this file])
Old Summary of Data on Amides in the Series
ELN post containing many of the raw Chemdraw and picture files for this wiki page - if you generate new pictures, add to the ELN page. This resource needs porting to Github.
Project Reports: Devon Scott and Eduvie Omene, University of Edinburgh, Tom MacDonald, The University of Sydney.
Online Meetings
Most recent online meeting (May 2016) relevant to this series. Recording.
This page is at http://tinyurl.com/OSM-Series4
Introduction
Preamble
The Triazolopyrazine (TP) Series, or Series 4, is the latest of the OSM series. It was announced by MMV and on the OSM blog (via the briefing document and as a general description) on September 10th 2013.

The series arises from work performed at Pfizer (that cannot be fully disclosed) which was followed by some hit-to-lead work funded directly by MMV and performed by a CRO (which can). The series includes many potent compounds, some with promising physicochemical properties, that are non-toxic and which clear assays such as hERG. Most importantly, members of the series have proven to be potent in vivo.
Current Series Aims
1. Lead optimisation, to improve solubility and metabolic stability while maintaining potency.
2. Validation of PfATP4 activity as mechanism of action via experimental and computational means.
3. To source more chemical and biological inputs from the widest possible community.
Notable Points about The Series
- Compounds in this series have been identified down to 16 nM potency.
- Seems to have good in vitro HLM and hHEP stability: CLint < 8.1 is compatible with 10 nM potency.
- RLM remains stubbornly high, particularly for the more potent analogues translating to short half-lives in rat PK. It's possible that rat metabolism may not be a good model for human metabolism for this series.
- Series appears to have little polypharmacology or cytotoxicity.
- There is strong correlation between activity vs. Pfal and activity in Kiaran Kirk’s ion regulation assay, implying the mechanism of action is inhibition of PfATP4.
Concerns about The Series
- Although dofetilide binding looks weak or nil, the series has shown activity in a patch clamp assay at Essen (1-10 μM) which is quite potent though with a window of >100 fold over Pfal potency.
- In Kip Guy’s resistant mutants the picture is mixed, but there is still support for the idea that some members of the series are weaker in the resistant strains. The series has no or weak >>1 μM activity against gametocytes, no activity against Winzeler’s Pb liver stage and may have weak activity against ookinetes but the dose-response data has not been completed.
Project Strands of Current Interest
The biggest issue is metabolic stability, as measured in rat in particular. There are few toxicity concerns. Thus possible future directions:
- Small scale changes around the side chains, particularly phenethyl to attempt to balance solubility, potency and metabolism. Other possibilities: a) N is tolerated in the pendant rings, but hasn’t been explored much recently. b) Is 3,4-diF the best substitution pattern? c) Some evidence (eg MMV669848) that the phenethyl side chain can be rigidified, perhaps the iso-indoline of that compound could be improved on with other ring systems and by more optimal substitution of the aromatic benzene ring of the isoindoline. d) The amide MMV670944 is interesting and shows good RLM stability, but many other amides failed to match its potency.
- Incorporation of a basic centre to increase volume as a potential fix for half-life. However, this might come at the expense of plasma concentration so would require high potency. Of the 29 compounds with a basic centre only one (MMV670437, below) has a measured potency < 100 nM (actually 44 nM).

- More significant structural changes. Of the changes made to the basic skeleton, the most successful might be the recent evaluation of the substitution position changes (e.g., MMV670945), possibly in combination with modifying the disposition of the N atoms in the core (though the triazolopyrazine has shown the best data, below).
SAR
Modification of Core Triazolopyrazine
To assess modification of the triazole part of the core, two compounds based on imadazopyrazines were made (MMV669846 and MMV670250). Both showed reduced potency against PfNF54 vs. the corresponding parent compound MMV639565. The RLM stability of MMV670250 was found to be poor.

The ring system in MMV669846 (imidazo[1,2-a]pyrazine) is also contained in the similar-looking Novartis series around compound KAI407, but the compounds apparently possess distinct mechanisms of action.
Modifications to the pyrazine part of the core were not well tolerated. Several aromatic variations were tried.

Several replacements of the aromatic ring with aliphatic rings were assayed.

As most of the analogs arising from core modifications were >1 μM potency, fewer were tested in RLM (though quite a few were tested in HLM). Of the 4 tested in RLM, the greatest stability had a Clint of 109, (HLM 9.5), several had HLM Clint 8 or less, particularly after moving or removing the N from the pyrazine ring. (<--need to locate primary data, since this disagrees with what is in the pictures here - MHT)
A selection of triazolopyridines were synthesised by Jamie Scott at The University of Sydney. Both ethers and amides were synthesised and found to be less potent than the corresponding trizaolopyrazine compounds.

Modification of Pyrazine Substitution Pattern
It was thought possible that the pyrazine moiety of the triazolopyrazine could undergo aldehyde oxidase-mediated metabolism at positions alpha- to the nitrogen. Four variations were made to the substitution of the "southernmost" (8-) ring C-H, all lowering potency vs. PfNF54.

Side chain transposition was investigated. The side-chain on the pyrazine ring was shifted to the adjacent carbon. The chain length was varied (n= 0,1,2) and linked through either O or N. Among the ethers, the phenethyl ether MMV670945 showed good potency (Pfal IC50 34 nM) but a poor stability in RLM (Cl 100 mL/min/Kg).
Modification of the Triazole Substitution
Attempts at lowering the lipophilicity of the compounds by replacing the triazole aryl substituent with a cyclo(hetero)aliphatic group, linked either by the heteroatom or otherwise (e.g. piperidine, tetrahydropyran, indoline or isoindoline) lowered the potency against PfNF54, as did an aniline substituent. A dimethylpyrazole substituent was also deleterious - note the comparison with MMV670944, which is potent.
One of the original inherited documents for Series 4 indicated that "heteroaryl" was not tolerated in this position, but the data do not currently support this as a blanket conclusion. Pyridinyl has been found to be a poor substituent, but one inherited compound containing a substituted pyridine was active.

Where an aromatic ring has been used in this position, it appears that a para-OCF3 and -OCHF2 groups generate higher potency than -OCH3.

A selection of benzylic alcohols were synthesised containing different substitution on the triazole ring. Once again, a paraphenyl-OCHF2 group led to potency whereas interestingly, a phenyl substituent led to inactivity, 2-substituted pyridine displayed micromolar activity, again suggesting that substituted pyridines could be further explored. Both cyclic and acyclic aliphatic compounds were inactive.

Pyrazine Side Chain Modifications - Ethers
The phenethyl ether side-chain on the pyrazine ring was flagged as a metabolic hot-spot and so several strategies were adopted to mitigate this risk. Some trends:
- Changing the length of the side-chain to anything other than 3 atoms severely lowers potency against PfNF54
- The linker atom to the pyrazine ring is crucial (O>>C>N)
- Heteroatoms in the side-chain lowered potency with the exception of MMV669848 (not an ether - features an isoindolino-methyl group on the pyrazine ring - see later section) although this compound had poor RLM stability.
- Constraining the linear side-chain into ring systems (e.g, azetidines, pyrrolidines, pyrazoles) severely reduced potency.
A 2-naphthol substituent on the pyrazine ring showed reasonable potency against Pfal (IC50 114 nM) but suffered from poor RLM stability (what is the number?). Hetero-analogs of 2-naphthol e.g. indole, indazole, quinoline, chroman, benzisoxazole, quinazoline, exhibited reduced potency.
In a separate attempt to mitigate the potential metabolism in the ethyl chain it was replaced by an aromatic group through inclusion of a phenol substituent on the pyrazine ring. Some substituted phenolates were metabolically more stable in vitro as well as in vivo in rat, although with reduced potency. (what are the numbers?)
Considering that the benzylic position in the phenethyl side-chain is prone to metabolic oxidation, several compounds having mono- and di-substitution in the benzylic position were made. Di-substitution lowered the potency considerably whereas mono-substitution with OMe, OCHF2, CH2OH, NMe2 groups retained good potency. This pattern was confirmed with the synthesis of phenyl analogues (MMV6888-96, -98 and 99). Synthesis of the corresponding dimethylamine is ongoing. Additional substitution alpha- to the ether oxygen led to complete loss of potency. The alpha-OCHF2 compound MMV670652, with a p-CN-phenyl group on the triazole ring, showed better RLM stability (cLogP effect). (what is the number?
Chris Swain suggested the synthesis of some benzyl ethers. MMV675959 was found to possess moderate activity. More of these compounds may be synthesised in July/Aug 2015, pending online consultation. In particular, 2-, 3- and 4-chlorosubstituted benzene and alpha-substituted benzylic ethers are of interest.

Planning of ether analogs: GHI 174 and blog appeal. Planning of more analogs in July 2014: GHI 232.
Pyrazine Side Chain Modifications - Amides
A small library of amides (including the m-Cl benzylamide, MMV668958) showed promising potency. However, other amides (derived from aliphatic or anilines) either showed lower potency or were inactive. The RLM of MMV668958 was poor - perhaps due to benzylic oxidation. Alpha-substitution at the benzylic position or constraining the benzylamine into an aminoindane did not improve potency. Several attempts to make aniline-amides with improved potency against Pfal failed. Loading the aniline ring with lipophilic substituents marginally improved potency but led to poor RLM stability.
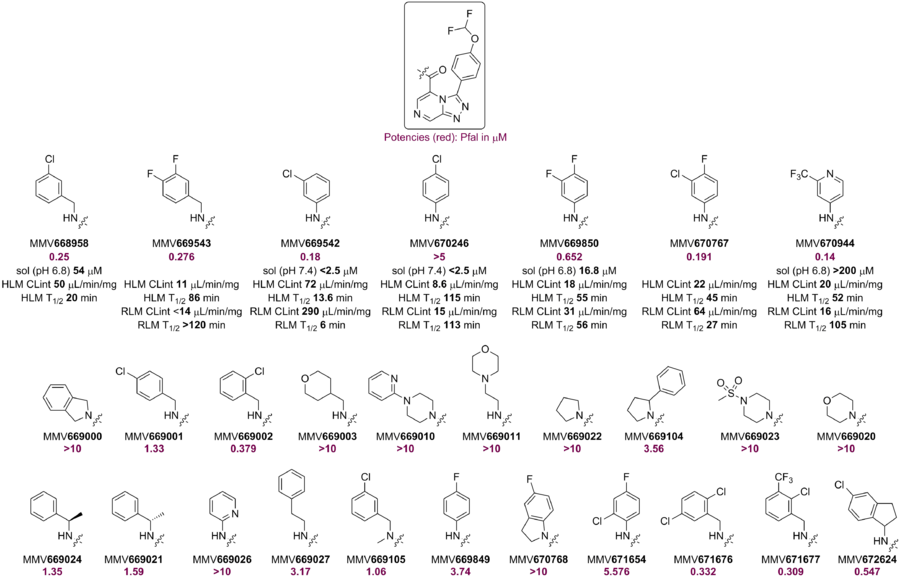
The p-Cl benzanilide MMV670246, although not active against PfNF54, showed good RLM stability perhaps due to lack of benzylic metabolism. However, its rat PK showed high clearance. The potent m-Cl (MMV669542) analog had poor RLM stability.

Planning of amide analogs: GH Issues 110, 101, 123, 207, Joie Garfunkel's suggestions. Planning of more amide analogs in July 2014: GHI 232.
Pyrazine Side Chain Modifications - Others

Vito Spadavecchio's potency predictor
This is a Linux based tool that uses regression to predict the potency of a arbitrary series 4 molecule: https://github.com/spadavec/OSM-ML-
Physicochem/metabolism/PK
The scheme below shows all the inherited met/PK data inherited, plus additional data for compounds more recently sent to Monash (green). (Associated discussion: GHI 107). Dec 2016: Tool for predicting sites of metabolism
For the more recent set, solubility was still seen as low and needs improving. The metabolic stability measured for the recent set appears not to be a mouse-specific event and needs to be improved to deliver a candidate. Met ID data (below) could help in the design of new analogs with improved stability. Is there a correlation between metabolic stability and Log D?
Some inherited data where the nature of the assay used was not clear: MMV669360 CLint 142 uL/min/kg and t1/2 = 12 min.
The PK curve for the relatively weak amide MMV670246 is shown below for oral & IV legs & parameters. The original data are contained in this picture.

A compound with better in vitro balance is MMV670652 but this compound has not been in rat PK. It may be possible to improve potency by synthesis of the more potent enantiomer - this is being addressed through synthesis of the synthetically simpler methyl analog OSM-S-208, the enantioenriched version of which was MMV669844.
The data for MMV639565 were contained in an inherited PDF summary of pharmacokinetics and efficacy.
As would be expected HLM vs. RLM shows a general correlation with approx 4-fold shift on average. However, for most of the more potent analogs, this increases to over 10-fold. The figure below shows the 4 sub 30 nM compounds with HLM & RLM measured: MMV670652, MMV670945, MMV670438 and MMV670947. (It looks like there are several other compounds here exhibiting good metabolic parameters for which we do not have data)

Metabolism ID
MMV669844, MMV669848 and MMV670936 were sent for ID of metabolites and data were obtained.

The data suggest that the core heterocyclic ring is subjected to an oxidation (though it is not clear the extent to which the compound is oxidized, only that certain peaks are present). The nature of the side chain appeared to have little influence on the results, suggesting alteration of the side chain is probably not the way to stop turnover, meaning it is likely that amides made in this series in the near future will also suffer from this problem. Nevertheless the efficacy was seen as sufficiently promising that amides remain attractive as they are. i.e. they are sufficiently robust for this point in the project. Residual question: what is the mechanism of the clearance (oxidation)? Is it CYP mediated? Open action item here. One possible route of metabolism is Aldehyde Oxidase, discussed in the next section.
Aldehyde Oxidase Assay
The possibility that the compounds were cleared by aldehyde oxidase was discussed (GHI 214), compounds were sent to Pfizer for evaluation and data returned from Scott Obach's laboratory (along with comments from him), summarized below, indicating that some Series 4 compounds were substrates for AO, and others are not.
For reference there is a very nice summary of AO over at Cambridge MedChem Consulting
Liver Stage
Two compounds have (MMV669844 and MMV670944, below) been evaluated vs. Pb liver schizonts and found to possess low activity vs. their blood stage potencies. Data here (along with a discussion of the similarity of Series 4 to the Novartis compound KAI407). (The original Series 4 briefing document stated the series had "no activity against Winzeler’s Pb liver stage" but there were no associated data to support this statement.)

Gametocyte Stage
The original Series 4 briefing document stated the series had "no or weak >>1uM activity against gametocytes" but there are no associated data to support this statement.
In Vivo Efficacy
MMV639565 and MMV669844 have both demonstrated parasite clearance in vivo, providing a highly attractive feature of this series.

The therapeutic efficacy of MMV639565 against P falciparum growing in peripheral blood of NODscidIL2Rγnull mice engrafted with human erythrocytes (po dosing, qd for 4 days) is shown below. A rapid parasite clearance - ED90 6.3 mg/kg was recorded.

Values in brackets indicate dose corrected according to quality control of formulation.
The data for MMV669844 show 99.9% inhibition of parasitemia in a snapshot (one dose level of 4 x 50 mg/kg).
Other Observations
Lipophilicity & lipophilic efficiency:
Few compounds achieve a lipophilic efficiency (as measured by pIC50 – AlogP) of greater than 4.0

Mechanism of Action: Possible PfATP4 Activity Deduced from Parasite Ion Regulation Assays
The following five compounds were evaluated in parasite ion regulation assays in the Kirk Laboratory; the hypothesis is that PfATP4 is a Na+ ATPase that exports Na+ and imports H+ (or equivalent) and that the effects of the compounds on Na+ concentration and pH are attributable to inhibition of this activity. Structures, potency, metabolism/solubility and raw PfATP4 assay data are here.

(Compounds that are inactive do not dissipate the plasma membrane Na+ gradient or increase the plasma membrane pH gradient, consistent with them not inhibiting PfATP4 at the concentration tested. The other compounds dissipated the plasma membrane Na+ gradient and increased the plasma membrane pH gradient at a concentration of 2 μM, consistent with them being PfATP4 inhibitors.)
There is a correlation: compounds inactive in these assays are not potent vs the parasite.
Testing was carried out vs. PfATP4-resistant mutants (ELN entry and GHI 251) in the laboratories of Kiaran Kirk and David Fidock data here. Also GHI251
PfATP4 is the apparent target of a number of different chemotypes.
PfATP4 is implicated in the MoA of several other leading antimalarials in development: the spiroindolone KAE609, the pyrazoleamides, the dihydroisoquinolone (+)-SJ-733 and various aminopyrazoles. A striking diversity of other compounds (from the malaria box) behave in the same way (summarised here), leading to the question: is PfATP4 really the target? There is no physical proof of binding between any of these compounds and PfATP4, because the protein has not yet been generated pure or crystallised. There is a homology model. Cross-resistance has been seen between parasites grown to be resistant to compound X (where there are mutations that are associated with PfATP4) and then tested with compound Y, including for the Series 4 compounds, as above. A current project strand is to develop a pharmacophore model that throws light on which compounds will be active in Kiaran's assay, and how they might be binding to PfATP4 (in part to allow us to de-prioritise the development of any more compounds having this same target). An initial attempt at developing this model was unsuccessful (i.e. not predictive - see the figure, where the "P Model predictions" correlate poorly with what was found in the ion regulation assay) possibly because the model did not allow for overlapping binding sites or take into consideration compound chirality. Model generation now needs to be re-attempted, and OSM needs community expertise in this area to proceed.
hERG Activity
MMV669844 and MMV670944 (OSM-S-175) were evaluated in a hERG assay, with the data indicating issues with both compounds that will need to be addressed. To investigate whether this is a problem for the series, several compounds (inherited and new, attempting to reduce cLogP - GH Issue #211) were evaluated for activity by AstraZeneca using this assay to see if this activity can be reduced. The results suggest that either the pendant OCHF2 or the amide is a problem. In contrast the previous two data points suggest that hERG activity can be obtained with compounds containing neither feature. Future analogs will need to address this. Cumulative data shown below (from here).

Former OSM postdoc Murray Robertson is applying literature hERG pharmacophore models to the series (GH Issue #188), and a community appeal is active for a laboratory willing to run higher-throughput binding assays (GH Issue #192). There is a lot of literature on hERG models (i.e. how to reduce hERG activity) with the most recent being an analysis of the ChEMBL database concluding that the dominant influence remains lipophilicity.
Toxicity
Toxicity has been evaluated at various points for this series and has generally been found to be low. e.g. here
Synthetic Chemistry
Synthetic Design
The approach to the synthesis of Series 4 up has mainly involved the functionalization of a pyrazine core, followed by a cyclization reaction to give the triazolopyrazine which may then be subsequently functionalized. The two main series, ether- and amide-linked compounds are shown below along with a specific example of a potent compound in each series.

Synthesis of the Ether-Linked Series
The synthesis of members of the ether series has been largely solved by Jo Ubels, as shown below.
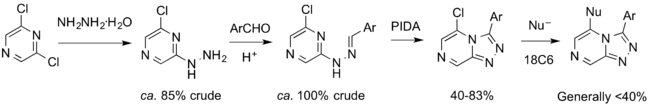
The cyclisation was achieved, initially not in the ca. 50% yields reported by the original CRO. Jo Ubels has solved this, with input from others such as a student working with Patrick Thomson, Devon Scott. The boys at Sydney Grammar School have also studied the early stages of the synthesis.
The union of the alcohol fragment with the chloro-intermediate was for a long time not proceeding cleanly, and this was also found by the CRO that first reported this route. Jo Ubels' lit survey found conditions involving a crown ether that has worked reliably. There have been periodic discussions about which alcohols to employ in the resulting synthetic scheme (post from April 2014, GHI174).
A synthetic challenge was the efficiency of the oxidative cyclization to form the triazolopyrazine. There was much discussion of the use of oxidants such as PIDA and chloramine T (Overview GHI 206) as well as the possibility that hydrazone isomers were cyclising at different rates (both Github Issue 97).
Some of the ether compounds contain a stereogenic centre in the benzylic position - structures are in the SAR section above, and the syntheses of these are dealt with below in the section of stereochemistry.
Synthesis of the Amide-Linked Series
Route 1: Initial Amide Bond Formation: It was decided (GHI121, GHI101) to pursue a route involving initial amide bond formation with the chloro-pyrazine acid shown below (OSM-S-150). This molecule is commercially available but is expensive, but there are now routes to it in the lab. Patrick and Inga planned this synthesis and then Inga completed it, as has Eduvie using the same approach (data missing in the lab notebook, but reports are available). [http://malaria.ourexperiment.org/triazolopyrazine_se/9189 Tom scaled up the Patrick/Eduvie/Inga approach and has been completed on a 15 g scale in four steps: 1 2 3 4. An assessment of the relative costs of synthesis vs. purchase was discussed (GHI163, GHI164) but not properly completed.

Sabin has successfully demonstrated an elegant alternative route to the starting pyrazine acid in one step from the methyl precursor (data).
For the subsequent coupling of the pyrazine acid with an amine, Tom and Alice have identified reliable conditions for the amide coupling using T3P that worked much better than an attempt to go via the acid chloride. There was an analysis of commercially-available primary and secondary amines that could be employed, the likely physical properties of the resulting compounds as well as an analysis of those amines available locally.
Hydrazone synthesis was surveyed by Inga) and is working reliably. (Discussion about making hydrazone first, then coupling with pyrazine: GHI152) Following work by Jo Ubels on optimizing the cyclization step for the ether series (above), hypervalent iodide PIDA may now be used to form the triazolopyrazine, as in this example.
Route 2: Carbonylation An efficient alternative synthetic approach would be to use the same chloro-triazolopyrazine core as for the ether series (above) and invent a new carbonylation method to introduce the amide as the final sequence (GHI101, GHI126, Inga's lit survey, Alice's original appeal for a collaborator, a summary of the associated discussion, Alice's framing of the relevant reactions (GHI205) and Tom's lit survey. The most recent results (GHI259) indicate there is a significant competing SNAr reaction when an alcohol was used as a nucleophile. The most recent reaction to date in this series was in Sept 2014.
Alternative Routes to the Triazolopyrazine Core
A route permitting more late-stage diversification of the core would involve synthesis of a halogenated core that could be subjected to cross-coupling or nucleophilic displacement. Two such routes are shown below.

Preliminary progress towards both has been made by Tom Macdonald (link to Honours thesis coming). The first steps (acid-mediated cyclizations) have been successfully carried out for Halogenation Routes 1 and 2. The bromination step in Route 1 has reportedly been carried out (we lack the experimental write-up) and the halogenation in Route 2 was reportedly carried out in 58% yield (most recent lab book entry of presumably different attempt, more recent attempt by Alice). In 2016, Haochuan Mao completed a synthesis of the chloro-bromo compound, including securing its crystal structure, building on earlier work by Tianyi Zheng. A Haverford College superlab project also looked into this route.
Fluoroalkene Isostere
Jo Ubels suggested a fluoroalkene isostere for the amide series. Jo pursued a simplified version of this chemistry with a pyridine ring in place of the pyrazine. The boronic ester (rather than the acids, which were predicted and found to be unstable, GHI250) was synthesized along with the bromofluoroalkene, but the coupling of these fragments has not yet been well explored. (Link to Ubels Honours thesis coming). Most recent experiment in this series.

Spectroscopy
Suggestion of the study of aryl-aryl interactions, with lit: GHI 232.
Chirality/Stereogenic Centres in This Series
Racemates/Resolutions: Frontrunner compound MMV670652 was evaluated/inherited as a racemate. There was interest in separating the enantiomers (GHI115, GHI111, GHI120. However, the difluoromethyl group makes the compound slightly challenging to synthesize (GHI119 and blog post). It is not clear that this group is needed - there are related structures that could be tried, and compounds lacking this group (and indeed the whole stereogenic centre) are still active as shown in the scheme. MMV669844 (containing CH3 rather than CHF2) was an inherited compound, and was reportedly synthesized using a Sharpless epoxidation on the precursor styrene, but there are no data on the enantiomeric excess of the product - it is shown as a single enantiomer in the original briefing document, so it is presumed (only) that the sample possessed an ee. Compound OSM-S-208 (GHI166), identical to MMV669844 save that it was synthesized as a racemate, was found to be reasonably potent and can therefore be used as a surrogate for MMV670652 for the separation of enantiomers in this series (GHI111). (Note there is ongoing discussion about whether compounds differing in ee, including where the ee is not known, should have different codes (GHI172)).

Alternatives: The CF3-substituted compound shown has yet to be made but has a prohibitively high cLogP; however the hydroxyl-substituted compound remains of interest (GHI178) and the des-F analog is under investigation (GHI235). MMV675948 (OSM-S-265) was found to be reasonably potent, providing another simple analog suitable for enantiomer separation. GHI236). Methoxy-substitured ether analogs lacking the Ph in the side chain are under investigation (GHI238, GHI239) though the only related inherited analog (MMV670762) is inactive.
Generally speaking all inherited compounds should be treated as racemates unless there is reason not to.
One approach to the synthesis of the chiral alcohol fragment needed for the racemic samples (e.g. of MMV670652) requires a source of cyanide.
Another approach to enantioenriched compounds in this series is through benzylic amines, either the known compounds MMV670437, MMV670763 and MMV671651 or analogs thereof (see GHI242, GHI241 and GHI240 respectively. (All three inherited amines were supposedly enantioenriched).

Enantioselective Approaches:
Alice Williamson proposed an enantioselective approach to the benzylic-functionalized ether compounds (GHI167).
Other Sources of Compounds Relevant to this Series
(Original community request)
The compound series appears to be novel/unusual (related: GHI142) with little similarity to compounds in PubChem/Surechem, though Chris Southan (in the previous link) found a related compound (CID 7118230) with reported antimalarial activity, with a structure similar to those described above for modification of the triazolopyrazine core. Jo Ubels also carried out a search and found few compounds similar to Series 4.
A similarity was noted between Series 4 compounds and a compound from Novartis likely active against vivax. Paul Willis suggested the mechanism of action of the Novartis compounds was likely PI4K - data and reasoning given in this lab book entry. Chris Southan modelled the OSM and Novartis compounds, which provides additional evidence for a different mechanism of action.
Desirable Compounds Not Yet Synthesised




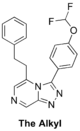




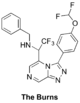
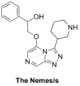
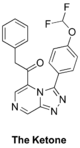
(Synthetic planning from Nov 2015 can be seen here)
Side chain derived from phenylalanine: GHI 254, ELN, OSM-S-261
ClC(C=C1)=CC=C1C2=NN=C3C=NC=C(OC[C@H](N)CC4=CC=CC=C4)N32 InChI=1S/C20H18ClN5O/c21-16-8-6-15(7-9-16)20-25-24-18-11-23-12-19(26(18)20)27-13-17(22)10-14-4-2-1-3-5-14/h1-9,11-12,17H,10,13,22H2/t17-/m1/s1
The Tianyi compound (contains methylated imidazopyrazine core, analogous to MMV669846): Discussion of Desirability: GHI358, GHI390 Haverford Planning OC(C1=CC=CC=C1)COC2=CN=CC3=NC(C)=C(C4=CC=C(OC(F)F)C=C4)N32 InChI=1S/C22H19F2N3O3/c1-14-21(16-7-9-17(10-8-16)30-22(23)24)27-19(26-14)11-25-12-20(27)29-13-18(28)15-5-3-2-4-6-15/h2-12,18,22,28H,13H2,1H3 VGNPLLBRBOUQAA-UHFFFAOYSA-N
The Heterooxazole (unexplored amide isostere in side chain): General discussion of the idea, Proposed synthesis, start of the synthesis, ELN FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(C4=CN=C(C5=CC=CC=C5)O4)N32 (free acid) InChI=1S/C21H13F2N5O2/c22-21(23)29-15-8-6-13(7-9-15)19-27-26-18-12-24-10-16(28(18)19)17-11-25-20(30-17)14-4-2-1-3-5-14/h1-12,21H FBTBZFMQUBZDSN-UHFFFAOYSA-N
The Heterooxadiazole (unexplored amide isostere in side chain): General discussion of the idea, Rationale, Proposed synthesis, Synthesis progress report by Haverford College, Relevant ELN, Revised synthetic route(s), Problems with imine formation, Discussion of potentially anomalous NMR spectrum, Second phase introduction and new ELN FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(C4=NC(C5=CC=CC=C5)=NO4)N32 InChI=1S/C20H12F2N6O2/c21-20(22)29-14-8-6-13(7-9-14)18-26-25-16-11-23-10-15(28(16)18)19-24-17(27-30-19)12-4-2-1-3-5-12/h1-11,20H HFPKPTYSFARKAA-UHFFFAOYSA-N
The hERG Evador/Acid Test (carboxylic acid analogue of MMV688896): Comment on desirability, Synthesis proposal, First synthesis attempt, summary and suggestions of ways forward with ELN, Discussion of synthetic roadblock, Discussion of Rationale. FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(OCC(C(O)=O)C4=CC=CC=C4)N32 InChI=1S/C21H16F2N4O4/c22-21(23)31-15-8-6-14(7-9-15)19-26-25-17-10-24-11-18(27(17)19)30-12-16(20(28)29)13-4-2-1-3-5-13/h1-11,16,21H,12H2,(H,28,29) FICNFVPYGUTYFV-UHFFFAOYSA-N
The Alkyl (Test to see if heteroatoms are needed on left-hand-side). Length may need tweaking - MMV669304 is potent): 1st proposed synthesis with commencement and ELN, 2nd proposed synthesis with ELN, FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(CCC4=CC=CC=C4)N32 InChI=1S/C20H16F2N4O/c21-20(22)27-17-10-7-15(8-11-17)19-25-24-18-13-23-12-16(26(18)19)9-6-14-4-2-1-3-5-14/h1-5,7-8,10-13,20H,6,9H2 QKTHSIJWBSHMDZ-UHFFFAOYSA-N
The Internal Alkyne (related to alkyl): Opening synthesis, ELN, discussion of rationale FC(F)OC1=CC=C(C2=NN=C3C=NC=C(C#CC4=CC=CC=C4)N32)C=C1 InChI=1S/C20H12F2N4O/c21-20(22)27-17-10-7-15(8-11-17)19-25-24-18-13-23-12-16(26(18)19)9-6-14-4-2-1-3-5-14/h1-5,7-8,10-13,20H KNVACGGMAGPRRW-UHFFFAOYSA-N
The Internal Alkene/Reduced Alkyl (synthetically related to alkyne and alkyl compounds; discussion of rationale of all three; The SMILES string is only for the cis isomer: Discussed, starting synthesis, Discussion of NMR spectrum of intermediate, ELN FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(/C=C\C4=CC=CC=C4)N32 InChI=1S/C20H14F2N4O/c21-20(22)27-17-10-7-15(8-11-17)19-25-24-18-13-23-12-16(26(18)19)9-6-14-4-2-1-3-5-14/h1-13,20H/b9-6- SGPCUHISMIPDDU-TWGQIWQCSA-N
The 6 (probe of tolerance to ring substitution): Synthesis proposal, Start of synthesis, ELN. Second phase Haverford attempt with second ELN FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC(C)=C(OCC(O)C4=CC=CC=C4)N32 InChI=1S/C21H18F2N4O3/c1-13-20(29-12-17(28)14-5-3-2-4-6-14)27-18(11-24-13)25-26-19(27)15-7-9-16(10-8-15)30-21(22)23/h2-11,17,21,28H,12H2,1H3 ZYFKQWHTACKGJR-UHFFFAOYSA-N
The 8 (also a probe of tolerance to ring substitution): Haverford synthesis plan, Start of Haverford synthesis, ELN, Rationale, Summary and closure of first attempt, cross-link to new attempt at 6- and 8- FC(F)OC(C=C1)=CC=C1C2=NN=C3C(C)=NC=C(OCC(O)C4=CC=CC=C4)N32 InChI=1S/C21H18F2N4O3/c1-13-19-25-26-20(15-7-9-16(10-8-15)30-21(22)23)27(19)18(11-24-13)29-12-17(28)14-5-3-2-4-6-14/h2-11,17,21,28H,12H2,1H3 ASWYMQOLUDKKDJ-UHFFFAOYSA-N
The Burns (amide isostere): Proposed synthesis from Haverford College, Start of synthesis, Discussion of results of intermediate steps here and here, ELN including summary of outcomes of first attempt. Start of second attempt with new ELN. FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(C(C(F)(F)F)NCC4=CC=CC=C4)N32 InChI=1S/C21H16F5N5O/c22-20(23)32-15-8-6-14(7-9-15)19-30-29-17-12-27-11-16(31(17)19)18(21(24,25)26)28-10-13-4-2-1-3-5-13/h1-9,11-12,18,20,28H,10H2 NYRGMZUFRGWPBF-UHFFFAOYSA-N
The Nemesis (deplanarisation of right-hand-side): Haverford College synthetic plan, Start of first Haverford synthesis, ELN, Second phase of synthesis with same ELN. OC(C1=CC=CC=C1)COC2=CN=CC3=NN=C(C4CNCCC4)N32 InChI=1S/C18H21N5O2/c24-15(13-5-2-1-3-6-13)12-25-17-11-20-10-16-21-22-18(23(16)17)14-7-4-8-19-9-14/h1-3,5-6,10-11,14-15,19,24H,4,7-9,12H2 SNDLBGDEOQPBOQ-UHFFFAOYSA-N
The Ketone (analog of amide linker): Haverford College proposed syntheses, Start of synthesis, ELN. Also "ketone linker" folder in Ed Tse's lab notebook. Second Haverford College campaign with new ELN. FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(C(CC4=CC=CC=C4)=O)N32 InChI=1S/C20H14F2N4O2/c21-20(22)28-15-8-6-14(7-9-15)19-25-24-18-12-23-11-16(26(18)19)17(27)10-13-4-2-1-3-5-13/h1-9,11-12,20H,10H2 KALFSUSYOZTBSP-UHFFFAOYSA-N
Strings for Google
Use this section to paste strings to make the page more discoverable.
Format (to allow creation of SDF): MMV Code (OSM Code and link to page) SMILES InChI InChIKey Potency in μmol
MMV639565 (OSM-S-272) FC1=C(F)C=CC(CCOC2=CN=CC3=NN=C(C4=CC=C(Cl)C=C4)N32)=C1 InChI=1S/C19H13ClF2N4O/c20-14-4-2-13(3-5-14)19-25-24-17-10-23-11-18(26(17)19)27-8-7-12-1-6-15(21)16(22)9-12/h1-6,9-11H,7-8H2 PMHUSEXABGDNGS-UHFFFAOYSA-N 0.038
MMV639725 (OSM-S-189) N#CC(C=C1)=CC=C1C2=NN=C3C=NC=C(OCCC4=CC=CC=C4Cl)N32 InChI=1S/C20H14ClN5O/c21-17-4-2-1-3-15(17)9-10-27-19-13-23-12-18-24-25-20(26(18)19)16-7-5-14(11-22)6-8-16/h1-8,12-13H,9-10H2 DQNFHRXLTAOFIL-UHFFFAOYSA-N
MMV668822 FC1=C(F)C=CC(CCOC2=C[N+]([O-])=CC3=NN=C(C4=CC=C(OC(F)F)C=C4)N32)=C1 InChI=1S/C20H14F4N4O3/c21-15-6-1-12(9-16(15)22)7-8-30-18-11-27(29)10-17-25-26-19(28(17)18)13-2-4-14(5-3-13)31-20(23)24/h1-6,9-11,20H,7-8H2
MMV668823 FC1=C(F)C=CC(CCOC2=CN=C(Cl)C3=NN=C(C4=CC=C(OC(F)F)C=C4)N32)=C1 InChI=1S/C20H13ClF4N4O2/c21-17-19-28-27-18(12-2-4-13(5-3-12)31-20(24)25)29(19)16(10-26-17)30-8-7-11-1-6-14(22)15(23)9-11/h1-6,9-10,20H,7-8H2
MMV668824 FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(CCO)N32 InChI=1S/C14H12F2N4O2/c15-14(16)22-11-3-1-9(2-4-11)13-19-18-12-8-17-7-10(5-6-21)20(12)13/h1-4,7-8,14,21H,5-6H2
MMV668955 FC(F)OC(C=C1)=CC=C1C2=NN=C3N2C(CCO)CNC3 InChI=1S/C14H16F2N4O2/c15-14(16)22-11-3-1-9(2-4-11)13-19-18-12-8-17-7-10(5-6-21)20(12)13/h1-4,10,14,17,21H,5-8H2
MMV668956 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OC4CN(C5=CC(F)=C(F)C=C5)C4 InChI=1S/C21H15F4N5O2/c22-16-6-3-13(7-17(16)23)29-10-15(11-29)31-19-9-26-8-18-27-28-20(30(18)19)12-1-4-14(5-2-12)32-21(24)25/h1-9,15,21H,10-11H2
MMV668957 FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(N4CCC(C5=CC=CC=C5)C4)N32 InChI=1S/C22H19F2N5O/c23-22(24)30-18-8-6-16(7-9-18)21-27-26-19-12-25-13-20(29(19)21)28-11-10-17(14-28)15-4-2-1-3-5-15/h1-9,12-13,17,22H,10-11,14H2
MMV668958 (OSM-S-176) FC(F)OC(C=C1)=CC=C1C2=NN=C3N2C(C(NCC4=CC(Cl)=CC=C4)=O)=CN=C3 InChI=1S/C20H14ClF2N5O2/c21-14-3-1-2-12(8-14)9-25-19(29)16-10-24-11-17-26-27-18(28(16)17)13-4-6-15(7-5-13)30-20(22)23/h1-8,10-11,20H,9H2,(H,25,29)
MMV668959 FC1=C(F)C=CC(CCOC2=CN=CC3=NN=C(C4CCOCC4)N32)=C1 InChI=1S/C18H18F2N4O2/c19-14-2-1-12(9-15(14)20)3-8-26-17-11-21-10-16-22-23-18(24(16)17)13-4-6-25-7-5-13/h1-2,9-11,13H,3-8H2
MMV668960 FC(F)OC(C=C1)=CC=C1C2=NN=C3N2CCNC3 InChI=1S/C12H12F2N4O/c13-12(14)19-9-3-1-8(2-4-9)11-17-16-10-7-15-5-6-18(10)11/h1-4,12,15H,5-7H2
MMV668961 FC1=C(F)C=CC(CCOC2=CN=CC3=NN=C(C4CCN(C(C)=O)CC4)N32)=C1 InChI=1S/C20H21F2N5O2/c1-13(28)26-7-4-15(5-8-26)20-25-24-18-11-23-12-19(27(18)20)29-9-6-14-2-3-16(21)17(22)10-14/h2-3,10-12,15H,4-9H2,1H3
MMV668962 FC(F)OC(C=C1)=CC=C1C2=NN=C3N2CCN(C(CN)=O)C3 InChI=1S/C14H15F2N5O2/c15-14(16)23-10-3-1-9(2-4-10)13-19-18-11-8-20(12(22)7-17)5-6-21(11)13/h1-4,14H,5-8,17H2
MMV669000 (OSM-S-177) O=C(N1CC(C=CC=C2)=C2C1)C3=CN=CC4=NN=C(C5=CC=C(OC(F)F)C=C5)N43 InChI=1S/C21H15F2N5O2/c22-21(23)30-16-7-5-13(6-8-16)19-26-25-18-10-24-9-17(28(18)19)20(29)27-11-14-3-1-2-4-15(14)12-27/h1-10,21H,11-12H2
MMV669001 (OSM-S-178) c1ncc2n(c1C(NCc1ccc(cc1)Cl)=O)c(nn2)c1ccc(cc1)OC(F)F InChI=1S/C20H14ClF2N5O2/c21-14-5-1-12(2-6-14)9-25-19(29)16-10-24-11-17-26-27-18(28(16)17)13-3-7-15(8-4-13)30-20(22)23/h1-8,10-11,20H,9H2,(H,25,29) InChI=1S/C20H14ClF2N5O2/c21-14-5-1-12(2-6-14)9-25-19(29)16-10-24-11-17-26-27-18(28(16)17)13-3-7-15(8-4-13)30-20(22)23/h1-8,10-11,20H,9H2,(H,25,29)
MMV669002 in amide scheme
MMV669003 (OSM-S-179) in amide scheme
MMV669006 FC1=C(F)C=CC(CCOC2=CN=CC3=NN=C(NC4=CC=C(F)C(F)=C4)N32)=C1 InChI=1S/C19H13F4N5O/c20-13-3-1-11(7-15(13)22)5-6-29-18-10-24-9-17-26-27-19(28(17)18)25-12-2-4-14(21)16(23)8-12/h1-4,7-10H,5-6H2,(H,25,27)
MMV669008 FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(N4CC(OC5=CC(F)=C(F)C=C5)C4)N32 InChI=1S/C21H15F4N5O2/c22-16-6-5-14(7-17(16)23)31-15-10-29(11-15)19-9-26-8-18-27-28-20(30(18)19)12-1-3-13(4-2-12)32-21(24)25/h1-9,15,21H,10-11H2
MMV669009 FC1=C(F)C=CC(CCOC2=CN=CC3=NN=C(N4CCCCC4)N32)=C1 InChI=1S/C18H19F2N5O/c19-14-5-4-13(10-15(14)20)6-9-26-17-12-21-11-16-22-23-18(25(16)17)24-7-2-1-3-8-24/h4-5,10-12H,1-3,6-9H2
MMV669010 (OSM-S-180) c1ncc2n(c1C(N1CCN(CC1)c1ncccc1)=O)c(nn2)c1ccc(cc1)OC(F)F InChI=1S/C23H22F2N8O2/c24-23(25)35-17-6-4-16(5-7-17)21-30-29-20-14-26-13-18(33(20)21)22(34)28-15-31-9-11-32(12-10-31)19-3-1-2-8-27-19/h1-8,13-14,23H,9-12,15H2,(H,28,34) UYWFTHNNGLIMAY-UHFFFAOYSA-N
MMV669011 (OSM-S-181) c1ncc2n(c1C(NCCN1CCOCC1)=O)c(nn2)c1ccc(cc1)OC(F)F InChI=1S/C19H20F2N6O3/c20-19(21)30-14-3-1-13(2-4-14)17-25-24-16-12-22-11-15(27(16)17)18(28)23-5-6-26-7-9-29-10-8-26/h1-4,11-12,19H,5-10H2,(H,23,28) LYBWZYLEKBKOOG-UHFFFAOYSA-N
MMV669020 (OSM-S-185) c1ncc2n(c1C(N1CCOCC1)=O)c(nn2)c1ccc(cc1)OC(F)F InChI=1S/C17H15F2N5O3/c18-17(19)27-12-3-1-11(2-4-12)15-22-21-14-10-20-9-13(24(14)15)16(25)23-5-7-26-8-6-23/h1-4,9-10,17H,5-8H2 LFRCBCKOIXZMLV-UHFFFAOYSA-N
MMV669021 c1ncc2n(c1C(N1CCCC1)=O)c(nn2)c1ccc(cc1)OC(F)F in amide scheme
MMV669022 (OSM-S-182) InChI=1S/C17H15F2N5O2/c18-17(19)26-12-5-3-11(4-6-12)15-22-21-14-10-20-9-13(24(14)15)16(25)23-7-1-2-8-23/h3-6,9-10,17H,1-2,7-8H2 HDIHMYGJUUDJKO-UHFFFAOYSA-N
MMV669023 (OSM-S-184) in amide scheme
MMV669024 (OSM-S-186) in amide scheme
MMV669025 FC1=C(F)C=CC(CCOC2=CNC(C3=NN=C(C4=CC=C(OC(F)F)C=C4)N32)=O)=C1 InChI=1S/C20H14F4N4O3/c21-14-6-1-11(9-15(14)22)7-8-30-16-10-25-19(29)18-27-26-17(28(16)18)12-2-4-13(5-3-12)31-20(23)24/h1-6,9-10,20H,7-8H2,(H,25,29)
MMV669026 in amide scheme
MMV669027 in amide scheme
MMV669028 FC1=C(F)C=CC(CCOC2=CN=CC3=NN=C(N4CC(C=CC=C5)=C5C4)N32)=C1 InChI=1S/C21H17F2N5O/c22-17-6-5-14(9-18(17)23)7-8-29-20-11-24-10-19-25-26-21(28(19)20)27-12-15-3-1-2-4-16(15)13-27/h1-6,9-11H,7-8,12-13H2
MMV669102 FC1=C(F)C=CC(CCOC2=CN=C(N(CC)CC)C3=NN=C(C4=CC=C(OC(F)F)C=C4)N32)=C1 InChI=1S/C24H23F4N5O2/c1-3-32(4-2)22-23-31-30-21(16-6-8-17(9-7-16)35-24(27)28)33(23)20(14-29-22)34-12-11-15-5-10-18(25)19(26)13-15/h5-10,13-14,24H,3-4,11-12H2,1-2H3
MMV669103 FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(NS(CC4=CC=C(C(F)(F)F)C=C4)(=O)=O)N32 InChI=1S/C20H14F5N5O3S/c21-19(22)33-15-7-3-13(4-8-15)18-28-27-16-9-26-10-17(30(16)18)29-34(31,32)11-12-1-5-14(6-2-12)20(23,24)25/h1-10,19,29H,11H2
MMV669104 (OSM-S-183) in amide scheme
MMV669105 in amide scheme
MMV669304 FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(CCCC4=CC=CC=C4)N32 InChI=1S/C21H18F2N4O/c22-21(23)28-18-11-9-16(10-12-18)20-26-25-19-14-24-13-17(27(19)20)8-4-7-15-5-2-1-3-6-15/h1-3,5-6,9-14,21H,4,7-8H2
MMV669305 FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(CCN4CCOCC4)N32 InChI=1S/C18H19F2N5O2/c19-18(20)27-15-3-1-13(2-4-15)17-23-22-16-12-21-11-14(25(16)17)5-6-24-7-9-26-10-8-24/h1-4,11-12,18H,5-10H2
MMV669310 FC(F)OC(C=C1)=CC=C1C2=NN=C3N2C(CCCC4=CC=CC=C4)CNC3 InChI=1S/C21H22F2N4O/c22-21(23)28-18-11-9-16(10-12-18)20-26-25-19-14-24-13-17(27(19)20)8-4-7-15-5-2-1-3-6-15/h1-3,5-6,9-12,17,21,24H,4,7-8,13-14H2
MMV669311 FC1=C(F)C=CC(CCOC2=CN=C(N)C3=NN=C(C4=CC=C(OC(F)F)C=C4)N32)=C1 InChI=1S/C20H15F4N5O2/c21-14-6-1-11(9-15(14)22)7-8-30-16-10-26-17(25)19-28-27-18(29(16)19)12-2-4-13(5-3-12)31-20(23)24/h1-6,9-10,20H,7-8H2,(H2,25,26)
MMV669312 FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(CCN4CC(C=CC=C5)=C5C4)N32 InChI=1S/C22H19F2N5O/c23-22(24)30-19-7-5-15(6-8-19)21-27-26-20-12-25-11-18(29(20)21)9-10-28-13-16-3-1-2-4-17(16)14-28/h1-8,11-12,22H,9-10,13-14H2
MMV669353 FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(N4CC(C5=CC=CC=C5)C4)N32 InChI=1S/C21H17F2N5O/c22-21(23)29-17-8-6-15(7-9-17)20-26-25-18-10-24-11-19(28(18)20)27-12-16(13-27)14-4-2-1-3-5-14/h1-11,16,21H,12-13H2
MMV669360 FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(COCC4=CC=C(F)C(F)=C4)N32 InChI=1S/C20H14F4N4O2/c21-16-6-1-12(7-17(16)22)10-29-11-14-8-25-9-18-26-27-19(28(14)18)13-2-4-15(5-3-13)30-20(23)24/h1-9,20H,10-11H2
MMV669541 FC1=C(F)C=CC(CCOC2=CN=C(C)C3=NN=C(C4=CC=C(OC(F)F)C=C4)N32)=C1 InChI=1S/C21H16F4N4O2/c1-12-19-27-28-20(14-3-5-15(6-4-14)31-21(24)25)29(19)18(11-26-12)30-9-8-13-2-7-16(22)17(23)10-13/h2-7,10-11,21H,8-9H2,1H3
MMV669542 (OSM-S-202) FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(C(NC4=CC=CC(Cl)=C4)=O)N32 InChI=1S/C19H12ClF2N5O2/c20-12-2-1-3-13(8-12)24-18(28)15-9-23-10-16-25-26-17(27(15)16)11-4-6-14(7-5-11)29-19(21)22/h1-10,19H,(H,24,28)
MMV669543 in amide scheme
MMV669544 FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(CNC(C4=CC(Cl)=CC=C4)=O)N32 InChI=1S/C20H14ClF2N5O2/c21-14-3-1-2-13(8-14)19(29)25-10-15-9-24-11-17-26-27-18(28(15)17)12-4-6-16(7-5-12)30-20(22)23/h1-9,11,20H,10H2,(H,25,29)
MMV669784 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OC4=CC=C(Cl)C=C4 InChI=1S/C18H11ClF2N4O2/c19-12-3-7-13(8-4-12)26-16-10-22-9-15-23-24-17(25(15)16)11-1-5-14(6-2-11)27-18(20)21/h1-10,18H
MMV669844 (OSM-S-218) [C@H](COc1cncc2n1c(nn2)c1ccc(cc1)C#N)(c1cc(c(cc1)F)F)OC
MMV669846 (OSM-S-273) FC1=C(F)C=CC(CCOC2=CN=CC3=NC=C(C4=CC=C(Cl)C=C4)N32)=C1 InChI=1S/C20H14ClF2N3O/c21-15-4-2-14(3-5-15)18-10-25-19-11-24-12-20(26(18)19)27-8-7-13-1-6-16(22)17(23)9-13/h1-6,9-12H,7-8H2 MQHQNFQVEXYAMO-UHFFFAOYSA-N 0.110
MMV669848 FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(CN4CC(C=CC=C5)=C5C4)N32 InChI=1S/C21H17F2N5O/c22-21(23)29-18-7-5-14(6-8-18)20-26-25-19-10-24-9-17(28(19)20)13-27-11-15-3-1-2-4-16(15)12-27/h1-10,21H,11-13H2
MMV669849 in amide scheme
MMV669850 in amide scheme
MMV670243 FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(CCCC4=CC(F)=C(F)C=C4)N32 InChI=1S/C21H16F4N4O/c22-17-9-4-13(10-18(17)23)2-1-3-15-11-26-12-19-27-28-20(29(15)19)14-5-7-16(8-6-14)30-21(24)25/h4-12,21H,1-3H2
MMV670246 FC(F)OC(C=C1)=CC=C1C2=NN=C3N2C(C(NC4=CC=C(Cl)C=C4)=O)=CN=C3 InChI=1S/C19H12ClF2N5O2/c20-12-3-5-13(6-4-12)24-18(28)15-9-23-10-16-25-26-17(27(15)16)11-1-7-14(8-2-11)29-19(21)22/h1-10,19H,(H,24,28)
MMV670249 FC1=C(F)C=CC(CCOC2=CN=CC3=NN=C(N4CCC5=C4C=CC(F)=C5)N32)=C1 InChI=1S/C21H16F3N5O/c22-15-2-4-18-14(10-15)5-7-28(18)21-27-26-19-11-25-12-20(29(19)21)30-8-6-13-1-3-16(23)17(24)9-13/h1-4,9-12H,5-8H2
MMV670250 (OSM-S-274) FC1=C(F)C=CC(CCOC2=CN=CC3=CN=C(C4=CC=C(Cl)C=C4)N32)=C1 InChI=1S/C20H14ClF2N3O/c21-15-4-2-14(3-5-15)20-25-11-16-10-24-12-19(26(16)20)27-8-7-13-1-6-17(22)18(23)9-13/h1-6,9-12H,7-8H2 JHORUMGXLQTHAD-UHFFFAOYSA-N 0.830
MMV670437 FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(OC[C@H](N(C)C)C4=CC(F)=C(F)C=C4)N32 InChI=1S/C22H19F4N5O2/c1-30(2)18(14-5-8-16(23)17(24)9-14)12-32-20-11-27-10-19-28-29-21(31(19)20)13-3-6-15(7-4-13)33-22(25)26/h3-11,18,22H,12H2,1-2H3/t18-/m0/s1
MMV670438 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OCC4(COC4)C5=CC=C(F)C(F)=C5 InChI=1S/C22H16F4N4O3/c23-16-6-3-14(7-17(16)24)22(10-31-11-22)12-32-19-9-27-8-18-28-29-20(30(18)19)13-1-4-15(5-2-13)33-21(25)26/h1-9,21H,10-12H2
MMV670652 FC(F)OC(C1=CC(F)=C(F)C=C1)COC2=CN=CC3=NN=C(C4=CC=C(C#N)C=C4)N32 InChI=1S/C21H13F4N5O2/c22-15-6-5-14(7-16(15)23)17(32-21(24)25)11-31-19-10-27-9-18-28-29-20(30(18)19)13-3-1-12(8-26)2-4-13/h1-7,9-10,17,21H,11H2
MMV670659 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OC4=CC=C(C=CC=C5)C5=C4 InChI=1S/C22H14F2N4O2/c23-22(24)30-17-8-6-15(7-9-17)21-27-26-19-12-25-13-20(28(19)21)29-18-10-5-14-3-1-2-4-16(14)11-18/h1-13,22H
MMV670762 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OCC(F)(F)F InChI=1S/C14H9F5N4O2/c15-13(16)25-9-3-1-8(2-4-9)12-22-21-10-5-20-6-11(23(10)12)24-7-14(17,18)19/h1-6,13H,7H2
MMV670763 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OC[C@H](NC)C4=CC=C(F)C(F)=C4 InChI=1S/C21H17F4N5O2/c1-26-17(13-4-7-15(22)16(23)8-13)11-31-19-10-27-9-18-28-29-20(30(18)19)12-2-5-14(6-3-12)32-21(24)25/h2-10,17,21,26H,11H2,1H3/t17-/m0/s1
MMV670764 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OC4=CC=C(C(F)(F)F)C=C4 InChI=1S/C19H11F5N4O2/c20-18(21)30-14-5-1-11(2-6-14)17-27-26-15-9-25-10-16(28(15)17)29-13-7-3-12(4-8-13)19(22,23)24/h1-10,18H
MMV670765 FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(OC4=CC(Cl)=C(Cl)C=C4)N32 InChI=1S/C18H10Cl2F2N4O2/c19-13-6-5-12(7-14(13)20)27-16-9-23-8-15-24-25-17(26(15)16)10-1-3-11(4-2-10)28-18(21)22/h1-9,18H
MMV670767 in amide scheme
MMV670768 in amide scheme
MMV670934 FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(NC4=CC(Cl)=C(Cl)C=C4)N32 InChI=1S/C18H11Cl2F2N5O/c19-13-6-3-11(7-14(13)20)24-15-8-23-9-16-25-26-17(27(15)16)10-1-4-12(5-2-10)28-18(21)22/h1-9,18,24H
MMV670935 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OC4=CC=CC(Cl)=C4 InChI=1S/C18H11ClF2N4O2/c19-12-2-1-3-14(8-12)26-16-10-22-9-15-23-24-17(25(15)16)11-4-6-13(7-5-11)27-18(20)21/h1-10,18H
MMV670936 C(COc1cncc2n1c(nn2)c1cnc(cc1)C(F)(F)F)c1cc(c(cc1)F)F
MMV670944 (OSM-S-175) FC(F)OC(C=C1)=CC=C1C2=NN=C3N2C(C(NC4=CC=NC(F)=C4)=O)=CN=C3 InChI=1S/C18H11F3N6O2/c19-14-7-11(5-6-23-14)24-17(28)13-8-22-9-15-25-26-16(27(13)15)10-1-3-12(4-2-10)29-18(20)21/h1-9,18H,(H,23,24,28)
MMV670945 FC(F)OC(C=C1)=CC=C1C2=NN=C3N2C=C(OCCC4=CC=C(F)C(F)=C4)N=C3 InChI=1S/C20H14F4N4O2/c21-15-6-1-12(9-16(15)22)7-8-29-18-11-28-17(10-25-18)26-27-19(28)13-2-4-14(5-3-13)30-20(23)24/h1-6,9-11,20H,7-8H2
MMV670946 FC(F)OC(C=C1)=CC=C1C2=NN=C3N2C=C(OCC4=CC=C(F)C(F)=C4)N=C3 InChI=1S/C19H12F4N4O2/c20-14-6-1-11(7-15(14)21)10-28-17-9-27-16(8-24-17)25-26-18(27)12-2-4-13(5-3-12)29-19(22)23/h1-9,19H,10H2
MMV670947 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OCC(CO)C4=CC=C(F)C(F)=C4 InChI=1S/C21H16F4N4O3/c22-16-6-3-13(7-17(16)23)14(10-30)11-31-19-9-26-8-18-27-28-20(29(18)19)12-1-4-15(5-2-12)32-21(24)25/h1-9,14,21,30H,10-11H2
MMV670949 FC(F)OC(C=C1)=CC=C1C2=NN=C3N2C=C(NCC4=CC=C(F)C(F)=C4)N=C3 InChI=1S/C19H13F4N5O/c20-14-6-1-11(7-15(14)21)8-24-16-10-28-17(9-25-16)26-27-18(28)12-2-4-13(5-3-12)29-19(22)23/h1-7,9-10,19,24H,8H2
MMV671647 FC(C(F)=C1)=CC=C1[C@@H](N2CCOCC2)COC3=CN=CC4=NN=C(C5=CC=C(OC(F)F)C=C5)N43 InChI=1S/C24H21F4N5O3/c25-18-6-3-16(11-19(18)26)20(32-7-9-34-10-8-32)14-35-22-13-29-12-21-30-31-23(33(21)22)15-1-4-17(5-2-15)36-24(27)28/h1-6,11-13,20,24H,7-10,14H2/t20-/m0/s1
MMV671648 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OC4=CC=C(C(C)(C)C)C=C4 InChI=1S/C22H20F2N4O2/c1-22(2,3)15-6-10-16(11-7-15)29-19-13-25-12-18-26-27-20(28(18)19)14-4-8-17(9-5-14)30-21(23)24/h4-13,21H,1-3H3
MMV671649 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OC4=CC=C(CCCC5)C5=C4 InChI=1S/C22H18F2N4O2/c23-22(24)30-17-8-6-15(7-9-17)21-27-26-19-12-25-13-20(28(19)21)29-18-10-5-14-3-1-2-4-16(14)11-18/h5-13,22H,1-4H2
MMV671650 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OC4=CC(C(F)(F)F)=C(Cl)C=C4 InChI=1S/C19H10ClF5N4O2/c20-14-6-5-12(7-13(14)19(23,24)25)30-16-9-26-8-15-27-28-17(29(15)16)10-1-3-11(4-2-10)31-18(21)22/h1-9,18H
MMV671651 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OC[C@H](N)C4=CC=C(F)C(F)=C4 InChI=1S/C20H15F4N5O2/c21-14-6-3-12(7-15(14)22)16(25)10-30-18-9-26-8-17-27-28-19(29(17)18)11-1-4-13(5-2-11)31-20(23)24/h1-9,16,20H,10,25H2/t16-/m0/s1
MMV671652 FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(N[C@H](C4=CC=C(F)C(F)=C4)CO)N32 InChI=1S/C20H15F4N5O2/c21-14-6-3-12(7-15(14)22)16(10-30)26-17-8-25-9-18-27-28-19(29(17)18)11-1-4-13(5-2-11)31-20(23)24/h1-9,16,20,26,30H,10H2/t16-/m0/s1
MMV671653 FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(N4N=CC(C5=CC(F)=C(F)C=C5)=C4)N32 InChI=1S/C21H12F4N6O/c22-16-6-3-13(7-17(16)23)14-8-27-30(11-14)19-10-26-9-18-28-29-20(31(18)19)12-1-4-15(5-2-12)32-21(24)25/h1-11,21H
MMV671654 in amide scheme
MMV671655 FC(F)OC(C=C1)=CC=C1C2=NN=C3N2C=C(NC4=CC=C(F)C(F)=C4)N=C3 InChI=1S/C18H11F4N5O/c19-13-6-3-11(7-14(13)20)24-15-9-27-16(8-23-15)25-26-17(27)10-1-4-12(5-2-10)28-18(21)22/h1-9,18,24H
MMV671676 in amide scheme
MMV671677 in amide scheme
MMV671678 FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(N4C=CN=C4CC5=CC(F)=C(F)C=C5)N32 InChI=1S/C22H14F4N6O/c23-16-6-1-13(9-17(16)24)10-18-28-7-8-31(18)20-12-27-11-19-29-30-21(32(19)20)14-2-4-15(5-3-14)33-22(25)26/h1-9,11-12,22H,10H2
MMV671679 CC1=NN(C2=CC=C(F)C=C2)C(C)=C1C3=NN=C4C=NC=C(OCC(F)(F)F)N43 InChI=1S/C18H14F4N6O/c1-10-16(11(2)28(26-10)13-5-3-12(19)4-6-13)17-25-24-14-7-23-8-15(27(14)17)29-9-18(20,21)22/h3-8H,9H2,1-2H3
MMV671680 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OC4=CC=C(S(F)(F)(F)(F)F)C=C4 InChI=1S/C18H11F7N4O2S/c19-18(20)31-13-3-1-11(2-4-13)17-28-27-15-9-26-10-16(29(15)17)30-12-5-7-14(8-6-12)32(21,22,23,24)25/h1-10,18H
MMV671926 FC(F)OC(C=C1)=CC=C1C2=NN=C3N2C=C(OC4=CC=C(F)C(F)=C4)N=C3 InChI=1S/C18H10F4N4O2/c19-13-6-5-12(7-14(13)20)27-16-9-26-15(8-23-16)24-25-17(26)10-1-3-11(4-2-10)28-18(21)22/h1-9,18H
MMV671927 FC(F)OC(C=C1)=CC=C1C2=NN=C3N2C=C(NCCC4=CC=C(F)C(F)=C4)N=C3 InChI=1S/C20H15F4N5O/c21-15-6-1-12(9-16(15)22)7-8-25-17-11-29-18(10-26-17)27-28-19(29)13-2-4-14(5-3-13)30-20(23)24/h1-6,9-11,20,25H,7-8H2
MMV671929 CC1=NN(C2=CC=C(F)C=C2)C(C)=C1C3=NN=C4C=NC=C(OC5=CC=C(Cl)C(Cl)=C5)N43 InChI=1S/C22H15Cl2FN6O/c1-12-21(13(2)31(29-12)15-5-3-14(25)4-6-15)22-28-27-19-10-26-11-20(30(19)22)32-16-7-8-17(23)18(24)9-16/h3-11H,1-2H3
MMV671933 FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(O[C@H]4CN(C5CC5)[C@H]4C6=CC(F)=C(F)C=C6)N32 InChI=1S/C24H19F4N5O2/c25-17-8-3-14(9-18(17)26)22-19(12-32(22)15-4-5-15)35-21-11-29-10-20-30-31-23(33(20)21)13-1-6-16(7-2-13)34-24(27)28/h1-3,6-11,15,19,22,24H,4-5,12H2/t19-,22-/m0/s1
MMV671934 FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(O[C@H]4C(N(C5CC5)[C@H]4C6=CC(F)=C(F)C=C6)=O)N32 InChI=1S/C24H17F4N5O3/c25-16-8-3-13(9-17(16)26)20-21(23(34)32(20)14-4-5-14)36-19-11-29-10-18-30-31-22(33(18)19)12-1-6-15(7-2-12)35-24(27)28/h1-3,6-11,14,20-21,24H,4-5H2/t20-,21+/m0/s1
MMV672618 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OC4=CC(C=CN5C)=C5C=C4 InChI=1S/C21H15F2N5O2/c1-27-9-8-14-10-16(6-7-17(14)27)29-19-12-24-11-18-25-26-20(28(18)19)13-2-4-15(5-3-13)30-21(22)23/h2-12,21H,1H3
MMV672619 FC(F)OC(C=C1)=CC=C1C2=NN=C3N2C=C(O[C@@H]4C(N(C5CC5)[C@@H]4C6=CC(F)=C(F)C=C6)=O)N=C3 InChI=1S/C24H17F4N5O3/c25-16-8-3-13(9-17(16)26)20-21(23(34)33(20)14-4-5-14)36-19-11-32-18(10-29-19)30-31-22(32)12-1-6-15(7-2-12)35-24(27)28/h1-3,6-11,14,20-21,24H,4-5H2/t20-,21+/m1/s1
MMV672620 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OC4=CC=C(C=CC(C(F)(F)F)=N5)C5=C4 InChI=1S/C22H12F5N5O2/c23-21(24)34-14-5-2-13(3-6-14)20-31-30-18-10-28-11-19(32(18)20)33-15-7-1-12-4-8-17(22(25,26)27)29-16(12)9-15/h1-11,21H
MMV672621 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OC4=CC=C(C(C)=NO5)C5=C4 InChI=1S/C20H13F2N5O3/c1-11-15-7-6-14(8-16(15)30-26-11)28-18-10-23-9-17-24-25-19(27(17)18)12-2-4-13(5-3-12)29-20(21)22/h2-10,20H,1H3
MMV672622 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OC4=CC=C(C(C)=CC(C)(C)O5)C5=C4 InChI=1S/C24H20F2N4O3/c1-14-11-24(2,3)33-19-10-17(8-9-18(14)19)31-21-13-27-12-20-28-29-22(30(20)21)15-4-6-16(7-5-15)32-23(25)26/h4-13,23H,1-3H3
MMV672623 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OC4=CC=C(C=NN5C)C5=C4 InChI=1S/C20H14F2N6O2/c1-27-16-8-15(7-4-13(16)9-24-27)29-18-11-23-10-17-25-26-19(28(17)18)12-2-5-14(6-3-12)30-20(21)22/h2-11,20H,1H3
MMV672624 ClC(C=C1)=CC2=C1C(CC2)NC(C3=CN=CC4=NN=C(C5=CC=C(OC(F)F)C=C5)N43)=O InChI=1S/C22H16ClF2N5O2/c23-14-4-7-16-13(9-14)3-8-17(16)27-21(31)18-10-26-11-19-28-29-20(30(18)19)12-1-5-15(6-2-12)32-22(24)25/h1-2,4-7,9-11,17,22H,3,8H2,(H,27,31)
MMV672625 CC(N(C1=CC=C(F)C=C1)N=C2C)=C2C3=NN=C4C=NC=C(C(NC5=CC(C(F)(F)F)=NC=C5)=O)N43 InChI=1S/C23H16F4N8O/c1-12-20(13(2)35(33-12)16-5-3-14(24)4-6-16)21-32-31-19-11-28-10-17(34(19)21)22(36)30-15-7-8-29-18(9-15)23(25,26)27/h3-11H,1-2H3,(H,29,30,36)
MMV672626 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OC(C(F)(F)F)C4(CC4)C5=CC=C(F)C(F)=C5 InChI=1S/C23H15F7N4O2/c24-15-6-3-13(9-16(15)25)22(7-8-22)20(23(28,29)30)36-18-11-31-10-17-32-33-19(34(17)18)12-1-4-14(5-2-12)35-21(26)27/h1-6,9-11,20-21H,7-8H2
MMV672686 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OC4=CC5=NN(C)C=C5C=C4 InChI=1S/C20H14F2N6O2/c1-27-11-13-4-7-15(8-16(13)26-27)29-18-10-23-9-17-24-25-19(28(17)18)12-2-5-14(6-3-12)30-20(21)22/h2-11,20H,1H3
MMV672687 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OCC(O)C4=CC=C(F)C(F)=C4 InChI=1S/C20H14F4N4O3/c21-14-6-3-12(7-15(14)22)16(29)10-30-18-9-25-8-17-26-27-19(28(17)18)11-1-4-13(5-2-11)31-20(23)24/h1-9,16,20,29H,10H2
MMV672688 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OCC4=CC=CC5=C4C=CC=C5 InChI=1S/C23H16F2N4O2/c24-23(25)31-18-10-8-16(9-11-18)22-28-27-20-12-26-13-21(29(20)22)30-14-17-6-3-5-15-4-1-2-7-19(15)17/h1-13,23H,14H2
MMV672689 FC(F)OC(C=C1)=CC=C1C2=NN=C3C=NC=C(O[C@H]4CN(C5CC5)[C@@H]4C6=CC(F)=C(F)C=C6)N32 InChI=1S/C24H19F4N5O2/c25-17-8-3-14(9-18(17)26)22-19(12-32(22)15-4-5-15)35-21-11-29-10-20-30-31-23(33(20)21)13-1-6-16(7-2-13)34-24(27)28/h1-3,6-11,15,19,22,24H,4-5,12H2/t19-,22+/m0/s1
MMV672723 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OCC(O)(C)C4=CC=C(F)C(F)=C4 InChI=1S/C21H16F4N4O3/c1-21(30,13-4-7-15(22)16(23)8-13)11-31-18-10-26-9-17-27-28-19(29(17)18)12-2-5-14(6-3-12)32-20(24)25/h2-10,20,30H,11H2,1H3
MMV672725 (mentioned in Jo Ubels Ether scheme)
MMV672726 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OCC4CCC5=C4C=CC(Cl)=C5 InChI=1S/C22H17ClF2N4O2/c23-16-5-8-18-14(9-16)1-2-15(18)12-30-20-11-26-10-19-27-28-21(29(19)20)13-3-6-17(7-4-13)31-22(24)25/h3-11,15,22H,1-2,12H2
MMV672727 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OCC(F)(C)C4=CC=C(F)C(F)=C4 InChI=1S/C21H15F5N4O2/c1-21(26,13-4-7-15(22)16(23)8-13)11-31-18-10-27-9-17-28-29-19(30(17)18)12-2-5-14(6-3-12)32-20(24)25/h2-10,20H,11H2,1H3
MMV672730 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OC4=CC=C(N=C(C(F)(F)F)N=C5)C5=C4 InChI=1S/C21H11F5N6O2/c22-20(23)34-13-3-1-11(2-4-13)18-31-30-16-9-27-10-17(32(16)18)33-14-5-6-15-12(7-14)8-28-19(29-15)21(24,25)26/h1-10,20H
MMV672936 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OCC(F)C4=CC=C(F)C(F)=C4 InChI=1S/C20H13F5N4O2/c21-14-6-3-12(7-15(14)22)16(23)10-30-18-9-26-8-17-27-28-19(29(17)18)11-1-4-13(5-2-11)31-20(24)25/h1-9,16,20H,10H2
MMV672937 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OC4=CC=C(C=CS5)C5=C4 InChI=1S/C20H12F2N4O2S/c21-20(22)28-14-4-2-13(3-5-14)19-25-24-17-10-23-11-18(26(17)19)27-15-6-1-12-7-8-29-16(12)9-15/h1-11,20H
MMV672939 FC(F)OC(C=C1)=CC=C1C2=NN=C3C=CC(OCCC4=CC(F)=C(F)C=C4)=NN32 InChI=1S/C20H14F4N4O2/c21-15-6-1-12(11-16(15)22)9-10-29-18-8-7-17-25-26-19(28(17)27-18)13-2-4-14(5-3-13)30-20(23)24/h1-8,11,20H,9-10H2
MMV672940 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OC4=CC=C(C=C(F)C(F)=C5)C5=C4 InChI=1S/C22H12F4N4O2/c23-17-8-13-3-6-16(7-14(13)9-18(17)24)31-20-11-27-10-19-28-29-21(30(19)20)12-1-4-15(5-2-12)32-22(25)26/h1-11,22H
MMV672941 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OC4=CC=C(C(F)=CC(F)=C5)C5=C4 InChI=1S/C22H12F4N4O2/c23-14-7-13-8-16(5-6-17(13)18(24)9-14)31-20-11-27-10-19-28-29-21(30(19)20)12-1-3-15(4-2-12)32-22(25)26/h1-11,22H
MMV672942 FC(F)OC(C=C1)=CC=C1C2=NN=C3C=CC(NCCC4=CC=C(F)C=C4)=NN32 InChI=1S/C20H16F3N5O/c21-15-5-1-13(2-6-15)11-12-24-17-9-10-18-25-26-19(28(18)27-17)14-3-7-16(8-4-14)29-20(22)23/h1-10,20H,11-12H2,(H,24,27)
MMV672989 FC(F)OC(C=C1)=CC=C1C2=NN=C(N32)C=NC=C3OC4=CC=C(C=CC(OC)=N5)C5=C4 InChI=1S/C22H15F2N5O3/c1-30-19-9-5-13-2-8-16(10-17(13)26-19)31-20-12-25-11-18-27-28-21(29(18)20)14-3-6-15(7-4-14)32-22(23)24/h2-12,22H,1H3
MMV672990 FC(F)OC(C=C1)=CC=C1C2=NN=C3N2C=C(NC(CC4=CC=CC(Cl)=C4)=O)N=C3 InChI=1S/C20H14ClF2N5O2/c21-14-3-1-2-12(8-14)9-18(29)25-16-11-28-17(10-24-16)26-27-19(28)13-4-6-15(7-5-13)30-20(22)23/h1-8,10-11,20H,9H2,(H,25,29)
MMV672992 N#CC(C=C1)=CC=C1C2=NN=C3C=CC(NCCC4=CN=CC=C4)=NN32 InChI=1S/C19H15N7/c20-12-14-3-5-16(6-4-14)19-24-23-18-8-7-17(25-26(18)19)22-11-9-15-2-1-10-21-13-15/h1-8,10,13H,9,11H2,(H,22,25)
JS 11-1 ClC1=CC=CC2=NN=C(C3=CC=C(C#N)C=C3)N21 InChI=1S/C13H7ClN4/c14-11-2-1-3-12-16-17-13(18(11)12)10-6-4-9(8-15)5-7-10/h1-7H ROKQDRVNUIOPKS-UHFFFAOYSA-N >50
JS 19-1 ClC(C=CC=C1)=C1CCOC2=CC=CC3=NN=C(C4=CC=C(C#N)C=C4)N32 InChI=1S/C21H15ClN4O/c22-18-5-2-1-4-16(18)12-13-27-20-7-3-6-19-24-25-21(26(19)20)17-10-8-15(14-23)9-11-17/h1-11H,12-13H2 APTFJFHOJWNXBX-UHFFFAOYSA-N 5.2
JS 20-1 N#CC(C=C1)=CC=C1C2=NN=C3C=CC=C(OCCC4=CC=CC=C4)N32 InChI=1S/C21H16N4O/c22-15-17-9-11-18(12-10-17)21-24-23-19-7-4-8-20(25(19)21)26-14-13-16-5-2-1-3-6-16/h1-12H,13-14H2 PLXIJFIDAXQTSX-UHFFFAOYSA-N 6.8
JS 21-1 FC(C=C1)=C(F)C=C1CCOC2=CC=CC3=NN=C(C4=CC=C(C#N)C=C4)N32 InChI=1S/C21H14F2N4O/c22-17-9-6-14(12-18(17)23)10-11-28-20-3-1-2-19-25-26-21(27(19)20)16-7-4-15(13-24)5-8-16/h1-9,12H,10-11H2 JCMCKOQZSFBPIN-UHFFFAOYSA-N 3.5
JS 16-1 O=C(NC1=CC=NC(C(F)(F)F)=C1)C2=CC=CC3=NN=C(C4=CC=C(C#N)C=C4)N32 InChI=1S/C20H11F3N6O/c21-20(22,23)16-10-14(8-9-25-16)26-19(30)15-2-1-3-17-27-28-18(29(15)17)13-6-4-12(11-24)5-7-13/h1-10H,(H,25,26,30) PGPJMFWOQCDCCV-UHFFFAOYSA-N >50
MMV688896 OC(C1=CC=CC=C1)COC2=CN=CC3=NN=C(C4=CC=C(OC(F)F)C=C4)N32 InChI=1S/C20H16F2N4O3/c21-20(22)29-15-8-6-14(7-9-15)19-25-24-17-10-23-11-18(26(17)19)28-12-16(27)13-4-2-1-3-5-13/h1-11,16,20,27H,12H2 GEHCMLKWLSBHDX-UHFFFAOYSA-N 0.27
MMV688894 OC(C1=CC=CC=C1)COC2=CN=CC3=NN=C(C4=CC=CC=C4)N32 InChI=1S/C19H16N4O2/c24-16(14-7-3-1-4-8-14)13-25-18-12-20-11-17-21-22-19(23(17)18)15-9-5-2-6-10-15/h1-12,16,24H,13H2 HQWUAOFYEUBLBZ-UHFFFAOYSA-N >5
MMV688895 OC(C1=CC=CC=C1)COC2=CN=CC3=NN=C(C4=CN=C(C(F)(F)F)C=C4)N32 InChI=1S/C19H14F3N5O2/c20-19(21,22)15-7-6-13(8-24-15)18-26-25-16-9-23-10-17(27(16)18)29-11-14(28)12-4-2-1-3-5-12/h1-10,14,28H,11H2 YUTTVQUFLHMVCQ-UHFFFAOYSA-N 3.462, 3.665
MMV688892 OC(C1=CC=CC=C1)COC2=CN=CC3=NN=C(C4CCCCC4)N32 InChI=1S/C19H22N4O2/c24-16(14-7-3-1-4-8-14)13-25-18-12-20-11-17-21-22-19(23(17)18)15-9-5-2-6-10-15/h1,3-4,7-8,11-12,15-16,24H,2,5-6,9-10,13H2 LWDWZIVAQMSZHL-UHFFFAOYSA-N >5
MMV688893 CCC1=NN=C2N1C(OCC(O)C3=CC=CC=C3)=CN=C2 InChI=1S/C15H16N4O2/c1-2-13-17-18-14-8-16-9-15(19(13)14)21-10-12(20)11-6-4-3-5-7-11/h3-9,12,20H,2,10H2,1H3 ZMELTVBBQIUHNR-UHFFFAOYSA-N >10
MMV675718 (OSM-S-201)
MMV675719 (OSM-S-206)
MMV675946 (OSM-S-204)
MMV675947 (OSM-S-254)
MMV675948 (OSM-S-265)
MMV675949 (OSM-S-258)
MMV675950 (OSM-S-255)
MMV675951 (OSM-S-256)
MMV675952 (OSM-S-257)
MMV675959 (OSM-S-259) FC1=CC(COC2=CN=CC3=NN=C(C4=CC=C(C#N)C=C4)N32)=CC=C1F InChI=1S/C19H11F2N5O/c20-15-6-3-13(7-16(15)21)11-27-18-10-23-9-17-24-25-19(26(17)18)14-4-1-12(8-22)2-5-14/h1-7,9-10H,11H2 AUBWBFLGMHEXIB-UHFFFAOYSA-N 0.452
MMV675960 (OSM-S-260)
MMV675961 (OSM-S-262)
MMV675962 (OSM-S-263)
MMV675963 (OSM-S-271)
(OSM-S-187)
(OSM-S-188)
(OSM-S-189)
(OSM-S-190)
(OSM-S-191)
(OSM-S-203)
(OSM-S-205)
(OSM-S-207)
(OSM-S-208) racemate of OSM-S-218
(OSM-S-219)
(OSM-S-220)
