Molecular Recognition Laboratorium
Contact Information
Molecular Recognition Laboratorium,
Institute für medizinische Immunologie
CHARITÉ - UNIVERSITÄTSMEDIZIN BERLIN

Hessische Str. 3-4 D-10115 Berlin, Germany phone +49-30-450 524092 fax +49-30-450 524942 mail annette.hayungs@charite.de web charite.de
Group Leader
Group Members
- Bernhard Aÿ, Postdoc
- Prisca Boisguérin, Postdoc
- Zerrin Fidan, Doktorandin
- Annette Hayungs - Secretary
- Marc Hovestädt - Doktorand
- Ines Kretzschmar - CTA
- Christiane Landgraf - CTA
- Carsten Mahrenholz - Doktorand
- Judith Müller - Doktorandin
- Livia Otte - Postdoc
- Rolf-Dietrich Stigler - IT- u. Sicherheitsbeauftragter
- Víctor Tapia - Doktorand
- Julia Triebus - Diplomantin
- Lars Vouillème - Doktorand
- Eike Wolter - Diplomant
Research interest
Technological Development of the Peptide Array Technologies
by Victor Tapia
FIGURE
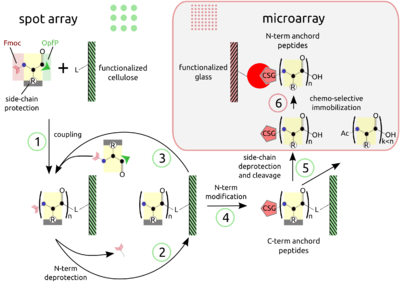
PEPTIDE ARRAYS The combination of SPOT peptide synthesis (figure A, steps 1 to 4) with
appropriate immobilization techniques on glass supports (figure A, steps
5 and 6) is wide spread. The SPOT technology provides low-scale but
high-throughput synthesis, while immobilization of pre-synthesized
peptides offers the benefit of a "chemical" purification step and
flexible array design. Additionally, the glass support is compatible
with fluorescence detection (see figure B, adapted from the web) and
offers the possibility to miniaturize binding assays. Beyond economy,
the later point is essential for quantitative measurements at the
steady-state of binding activity, as has been described [Ekins 1998] and
can be proven by the mass-action law.
The basic point of this technology is the simultaneous display of a
systematic collection of peptides on a planar support, on which numerous
bimolecular interaction assays can be carried out under homogeneous
conditions.
MORE
The technological advancement of peptide arrays can roughly be
reviewed by focusing (at least) on three milestones of stepwise peptide
synthesis. ( 1st ) The development of solid-phase peptide synthesis (SPPS) by
Bruce Merrifield [Gutte and Merrifield, 1969; Merrifield, 1965] and
adaptions of this procedure [Fields and Noble, 1990] set the chemical
ground for innovative technologies to follow. ( 2nd ) The development
of the “Pin” method by H. Geysen [Geysen, et al., 1984] introduces the
array format to peptide synthesis. ( 3rd ) Definitive establishment of peptide
arrays came along with the development of the SPOT synthesis by Roland
Frank [Frank, 1992; Frank, 2002] which simplified chemical synthesis of
peptide arrays to the addressable deposition of reagents on a cellulose
sheet. Moreover, chemical synthesis allows incorporation of non-natural
building blocks, preparation of branched and cyclic structures and
labelling with chromophores. Modern peptide synthesis approaches and
molecular biology make peptides accessible in a high degree of
structural diversity. The two greatest drawbacks of synthetic peptide
arrays are peptide length, with a quality threshold between 30 and 50
amino-acids, as well as the restriction to linear motives, since the
mimicry of nonlinear motives with linear peptide constructs is still
under development [Goede, et al., 2005].
Peptide arrays present also a
milestone in the advancement of analytical binding assay systems. Since
the 90s a major aspect of development to achieve the required
sensitivities to analyse biological samples has been the miniaturization
of analytical devices [Ekins, 1998]. It is important to note that
miniaturization is not only a matter of high-throughput and economy.
Miniaturization is an essential factor that should provide saturation of
binding sites under low analyte concentrations without significantly
altering its bulk (or ambient) concentration upon capturing [Ekins, et
al., 1990; Ekins, 1989; Joos, et al., 2002; Templin, et al., 2002]. In
this sense, the first application of a peptide microarray device in
1991, anticipating even the application of cDNA arrays, achieved already
the impressive feature density of about 1024 peptides in 1.6 cm2 by
means of in situ light-directed parallel synthesis [Fodor, et al.,
1991]. The several methods available to generate peptide arrays on
planar solid surfaces offer a range between 16 peptides per cm2, in the
case of SPOT macroarrays [Reimer, et al., 2002; Schutkowski, et al.,
2004], to 2000-4000 peptides in 1.5 cm2, in the case of microarrays
generated by digital photolithography [El Khoury, et al., 2007; Gao, et
al., 2004; Pellois, et al., 2000; Pellois, et al., 2002].
In order to
support synthesis, planar materials have to fulfil several requirements
including stability towards solvent and reagent deposition. The
functional groups on the surface must also be biochemically accessible
for chemical derivatisation. Furthermore, upon solid-phase binding
assay, generated peptides must be functionally displayed to allow
molecular recognition with a binding partner in the solution phase. In
particular non-specific interactions should be ruled out. Flexible
porous supports such as cellulose [Eichler, et al., 1989; Frank and
Döring, 1988], cotton [Eichler, et al., 1991; Schmidt and Eichler, 1993]
or membranes [Daniels, et al., 1989; Wang and Laursen, 1992; Wenschuh,
et al., 2000] are preferentially used for peptide array generation.
Rigid, non-porous materials such as glass [Falsey, et al., 2001], gold
films [Houseman and Mrksich, 2002; Jonsson, et al., 1991; Malmqvist,
1993], or silicon [Fodor, et al., 1991; Pellois, et al., 2002] have also
been used for in situ synthesis, but are much more technically
demanding. On the other side, rigid materials have a number of
advantages over porous supports for functional display of molecules.
Impermeability and smooth two-dimensionality of the material does not
limit diffusion of the binding partner and leads to more accurate
kinetics of recognition events. Finally the flatness and transparency of
glass improve image acquisition and simplifies the use of fluorescence
dyes for the read out process. In some cases, assembled 3D structure on
a non-porous surface could be a fruitful approach. Several techniques
for coherent surface modifications are described over the past twenty
years in the literature. For a comparative overview on this field we
refer to articles dedicated to the peptide and protein array
technologies [Angenendt and Glokler, 2004; Angenendt, et al., 2002;
Angenendt, et al., 2003; Seurynck-Servoss, et al., 2008;
Seurynck-Servoss, et al., 2007; Seurynck-Servoss, et al., 2007; Sobek,
et al., 2007; Sobek, et al., 2006; Wenschuh, et al., 2000].
REFERENCES
- Angenendt, P., and Glokler, J. (2004) Evaluation of antibodies and microarray coatings as a prerequisite for the generation of optimized antibody microarrays, Methods Mol Biol 264, 123-34.
- Angenendt, P., Glokler, J., Murphy, D., Lehrach, H., and Cahill, D. J. (2002) Toward optimized antibody microarrays: a comparison of current microarray support materials, Anal Biochem 309, 253-60.
- Angenendt, P., Glokler, J., Sobek, J., Lehrach, H., and Cahill, D. J. (2003) Next generation of protein microarray support materials: evaluation for protein and antibody microarray applications, J Chromatogr A 1009, 97-104.
- Daniels, S. B., Bernatowicz, M. S., Coull, J. M., and Köster, H. (1989) Membranes as solid supports for peptide synthesis., Tetrahedron Lett. 30.
- Eichler, J., Beyermann, M., and Bienert, M. (1989) Application of cellulose paper as support in simultaneous solid phase peptide synthesis., Colect. Czech. Chem. Commun. 54, 1746-52.
- Eichler, J., Bienert, M., Stierandova, A., and Lebl, M. (1991) Evaluation of cotton as a carrier for solid-phase peptide synthesis., Peptide Res. 4, 296-307.
- Ekins, R., Chu, F., and Biggart, E. (1990) Multispot, multianalyte, immunoassay, Ann Biol Clin (Paris) 48, 655-66.
- Ekins, R. P. (1989) Multi-analyte immunoassay, J Pharm Biomed Anal 7, 155-68.
- Ekins, R. P. (1998) Ligand assays: from electrophoresis to miniaturized microarrays, Clin Chem 44, 2015-30.
- El Khoury, G., Laurenceau, E., Dugas, V., Chevolot, Y., Merieux, Y., Duclos, M. C., Souteyrand, E., Rigal, D., Wallach, J., and Cloarec, J. P. (2007) Acid deprotection of covalently immobilized peptide probes on glass slides for peptide microarrays, Conf Proc IEEE Eng Med Biol Soc 2007, 2242-6.
- Falsey, J. R., Renil, M., Park, S., Li, S., and Lam, K. S. (2001) Peptide and small molecule microarray for high throughput cell adhesion and functional assays, Bioconjug Chem 12, 346-53.
- Fields, G. B., and Noble, R. L. (1990) Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids, Int J Pept Protein Res 35, 161-214.
- Fodor, S. P., Read, J. L., Pirrung, M. C., Stryer, L., Lu, A. T., and Solas, D. (1991) Light-directed, spatially addressable parallel chemical synthesis, Science 251, 767-73.
- Frank, R. (1992) Spot-synthesis: an easy technique for the positionally addressable, parallel chemical synthesis on a membrane support, Tetrahedron, 9217-32.
- Frank, R. (2002) The SPOT-synthesis technique: Synthetic peptide arrays on membrane supports--principles and applications, J. Immunol. Methods 267, 13-26.
- Frank, R., and Döring, R. (1988) Simultaneous multiple peptide synthesis under continuous flow conditions on cellulose paper disks as segmental solid supports, Tetrahedron 44, 6031-40.
- Gao, X., Pellois, J. P., Na, Y., Kim, Y., Gulari, E., and Zhou, X. (2004) High density peptide microarrays. In situ synthesis and applications, Mol Divers 8, 177-87.
- Geysen, H. M., Meloen, R. H., and Barteling, S. J. (1984) Use of peptide synthesis to probe viral antigens for epitopes to a resolution of a single amino acid, Proc Natl Acad Sci U S A 81, 3998-4002.
- Goede, A., Jaeger, I. S., and Preissner, R. (2005) SUPERFICIAL--surface mapping of proteins via structure-based peptide library design, BMC Bioinformatics 6, 223.
- Gutte, B., and Merrifield, R. B. (1969) The total synthesis of an enzyme with ribonuclease A activity, J Am Chem Soc 91, 501-2.
- Houseman, B. T., and Mrksich, M. (2002) Towards quantitative assays with peptide chips: a surface engineering approach, Trends Biotechnol 20, 279-81.
- Jonsson, U., Fagerstam, L., Ivarsson, B., Johnsson, B., Karlsson, R., Lundh, K., Lofas, S., Persson, B., Roos, H., Ronnberg, I., and et al. (1991) Real-time biospecific interaction analysis using surface plasmon resonance and a sensor chip technology, Biotechniques 11, 620-7.
- Joos, T. O., Stoll, D., and Templin, M. F. (2002) Miniaturised multiplexed immunoassays, Curr Opin Chem Biol 6, 76-80.
- Malmqvist, M. (1993) Biospecific interaction analysis using biosensor technology, Nature 361, 186-7.
- Merrifield, R. B. (1965) Automated synthesis of peptides, Science 150, 178-85.
- Pellois, J. P., Wang, W., and Gao, X. (2000) Peptide synthesis based on t-Boc chemistry and solution photogenerated acids, J Comb Chem 2, 355-60.
- Pellois, J. P., Zhou, X., Srivannavit, O., Zhou, T., Gulari, E., and Gao, X. (2002) Individually addressable parallel peptide synthesis on microchips, Nat Biotechnol 20, 922-6.
- Reimer, U., Reineke, U., and Schneider-Mergener, J. (2002) Peptide arrays: from macro to micro, Curr Opin Biotechnol 13, 315-20.
- Schmidt, M., and Eichler, J. (1993) Multiple peptide synthesis using cellulose-based carriers: Synthesis of substance P - diastereoisomers and their histamine-releasing activity., Bioorg. Med. Chem. Lett. 3, 441-46.
- Schutkowski, M., Reimer, U., Panse, S., Dong, L., Lizcano, J. M., Alessi, D. R., and Schneider-Mergener, J. (2004) High-content peptide microarrays for deciphering kinase specificity and biology, Angew Chem Int Ed Engl 43, 2671-4.
- Seurynck-Servoss, S. L., Baird, C. L., Miller, K. D., Pefaur, N. B., Gonzalez, R. M., Apiyo, D. O., Engelmann, H. E., Srivastava, S., Kagan, J., Rodland, K. D., and Zangar, R. C. (2008) Immobilization strategies for single-chain antibody microarrays, Proteomics 8, 2199-210.
- Seurynck-Servoss, S. L., Baird, C. L., Rodland, K. D., and Zangar, R. C. (2007) Surface chemistries for antibody microarrays, Front Biosci 12, 3956-64.
- Seurynck-Servoss, S. L., White, A. M., Baird, C. L., Rodland, K. D., and Zangar, R. C. (2007) Evaluation of surface chemistries for antibody microarrays, Anal Biochem 371, 105-15.
- Sobek, J., Aquino, C., and Schlapbach, R. (2007) Quality considerations and selection of surface chemistry for glass-based DNA, peptide, antibody, carbohydrate, and small molecule microarrays, Methods Mol Biol 382, 17-31.
- Sobek, J., Bartscherer, K., Jacob, A., Hoheisel, J. D., and Angenendt, P. (2006) Microarray technology as a universal tool for high-throughput analysis of biological systems, Comb Chem High Throughput Screen 9, 365-80.
- Templin, M. F., Stoll, D., Schrenk, M., Traub, P. C., Vohringer, C. F., and Joos, T. O. (2002) Protein microarray technology, Drug Discov Today 7, 815-22.
- Wang, Z., and Laursen, R. A. (1992) Multiple peptide synthesis on polypropylene membranes for rapid screening of bioactive peptides., Pep. Res. 5, 275-80.
- Wenschuh, H., Volkmer-Engert, R., Schmidt, M., Schulz, M., Schneider-Mergener, J., and Reineke, U. (2000) Coherent membrane supports for parallel microsynthesis and screening of bioactive peptides, Biopolymers 55, 188-206.
Structural Modularity in Protein-Protein Recognition
by Victor Tapia
FIGURE
Protein Interaction Domains
The organisation of living systems is a complex network of molecular
interactions. Proteins are a central component of the network as they
may bind to other proteins as well as to phospholipids, nucleic acids
and small molecules to interconnect the diverse physiological functions
of the cell. In the background of these observations, the existence of a
molecular recognition code for cellular organisation is very suggestive.
MORE TEXT
Structural analysis of functional protein complexes suggests at least
two classes of protein-protein interaction that may be extendable to the
other kinds of protein interactions. In the first class, the
complementary surfaces of the interacting partners are both extensive.
Under these circumstances, the residues involved in each interacting
surface come together only upon protein folding. The second class
consists on asymmetric interactions, where a protein domain (folding
motive of moderate size like a pocket on the protein’s surface) may dock
a short lineal peptide motive (a peptide ligand) on the partner protein.
While interactions over extensive surfaces cannot be inferred, the
binding determinants of a protein interaction domain (PID) may be mapped
to short peptides matching the sequence of the ligand peptide. The
importance of small recognition domains in the formation of protein
complexes by binding to short lineal peptides was demonstrated in the
late 1980s and early 1990s (Sadowski, I. et al., 1986; Ren, R. et al.,
1993; Mayer, B.J. et al., 1993).
In this era of extensive genome sequencing, many PIDs have been
discovered. The interaction partners and, therefore, the functions of
such proteins may be determined by identifying the critical binding sites for
one family member through evolutionary tracing (Lichtarge, O. et al., 1996) or
through high-parallel screening of functional protein arrays (Phizicky,
E. et al, 2003). Many of the PIDs in proteins can be grouped into
families that show clear evidence of their evolution from a common
ancestor, and genome sequences from Saccharomyces cerevisiae to Homo
sapiens reveal large numbers of proteins that contain one or more common
domains.
Src Homology Families - A PID Prototype
In a pioneering work on the kinase function and transforming activity of
the Fujinami Sarcoma Virus, Sadowski et al. (1986) discovered “a unique
domain… (which) is absent from kinases that span the plasma membrane”
and concluded that “the presence of this noncatalytic domain in all
known cytoplasmic tyrosine kinases of higher and lower eucaryotes argues
for an important biological function... the noncatalytic domain may
direct specific interactions of the enzymatic region with cellular
components that regulate or mediate tyrosine kinase function”. These
regions were called Src homology 2 (SH2) and 3 (SH3), the name SH1 being
reserved to the catalytic region. Since then, the gained knowledge on SH
domain function has been a paradigm in our understanding of PID
biochemistry.
MORE TEXT
The structure of SH2 family members involves about 100 residues that, in
the case of the kinase Src, are located N terminal to the catalytic
region and resembles a pocket dominated by a ß-sheet sandwiched between
a pair of a-helices. SH2 domains bind the protein containing them to a
second protein on a phosphorylated tyrosine residue (pY) in a specific
amino acid sequence context (Ladbury, J.E. et al., 2000).
The SH3 domain structure, also found in cytoplasmic kinases like Src,
consists largely of two ? sheets that form a partly open ß-barrel. The
ligand-binding site is a hydrophobic surface showing three shallow
pockets or grooves defined by conserved aromatic residues. The ligand
adopts an extended, left-handed helical conformation termed the
polyproline-2 (or PPII) helix. Two of the binding pockets of the SH3
domain are occupied by two hydrophobic proline dipeptides on two
adjacent turns of the helix, whereas the third ‘specificity’ pocket in
most cases interacts with a basic residue in the ligand distal to the
xPxxP core conserved motive of the PPII helix (Mayer, B.J. et al.,
2001).
The amino acids located at the binding site for the
phosphorylated polypeptide of SH2 and for the polyprolin core of SH3
have been the slowest to change during the long evolutionary process
that produced the large SH2 and SH3 families of peptide recognition
domains. Because mutation is a random process, this result is attributed
to the preferential elimination during evolution of all organisms whose
SH domains became altered in a way that inactivated the SH-binding site,
thereby destroying the function of the SH domain. Are the PID/ligand
interactions specific enough or must a certain interaction compete with
the bulk of structurally similar structures in a struggle for dynamical
complex formation?
From Promiscuous Recognition Events to Mutually Exclusive Cellular Responses
UPCOMMING: list of cited authors
The elucidation of functional pathways of signal transduction,
biochemical function or gene regulation, is firstly addressed in
proteomics by deriving interaction networks depicting ideally all
interactions in the cell. Several attempts have been done in this
direction on different model organisms and with varied methods,
including co-purification by affinity chromatography (Ho, Y. et al.,
2002; Gavin, A.-C. et al., 2002; Bouwmeester, T. et al., 2004), yeast
two-hybrid, phage display, peptide array technologies, etc. A comparison
of datasets derived by individual methods demonstrates that
different methods have different potential. For example, affinity
chromatographic approaches are biased to tight interactions such as
those involving extensive complementary surfaces, while
interactions in which one of the two partners contains at least one
PID are more frequent in the two-hybrid database. The higher
sensitivity of the so called synthetic approaches (yeast
two-hybrid, phage display and peptide array technologies) make them
better suited for detecting PID-mediated interactions since their
peptide affinity in terms of Kd falls in the 10 – 100 µM range (high Kd
--> low affinity). However, this advantage is counterbalanced by a low
specificity, especially of the yeast two-hybrid approach (Phizyki, 2002).
MORE TEXT
In order to correct this deficiency a double check-up of the information
fed into the interaction databases is recommended. This can be achieved
by deriving two interaction networks through orthogonal (fundamentally
different) synthetic methods and then considering only the intersection
between the two datasets (Tong, A.H.Y. et al.,2002; Castagnoli, L. et
al., 2004; Landgraf, C. et al., 2004). False positive reports are thus
reduced if the causes for measurement error are different in each
method.
The strength of this combined approach to deliver
physiologically relevant interactions has been proven for a phage
display/yeast two-hybrid intersected dataset (Tong, A.H.Y. et al.,2002).
A notable conclusion of this approach is that the intersected dataset of
proteins that are able to interact with a given PID is larger than
expected when cellular events are viewed as a precise wiring of the
proteins in the cell. Although a set of these biochemically potential
binders may have no physiological relevance due to expression at
different times or tissues, in vitro disrupted structures, etc., the
paradox of promiscuous recognition and mutually exclusive responses
seems to be inherent to PID mediated interactions: the recent work of
Landgraf et al. (2004) supports the observation that a large fraction of
natural peptides with the biochemical potential to bind to any given SH3
domain is actually used in vivo to mediate the formation of a
complex.
An additional difficulty to derive functional
interaction pathways is that the difference in affinity between
‘specific’ and ‘non-specific’ interactions has been shown to be less
than two orders of magnitude in the case of SH2 and its peptide ligands
(Songyang, Z. et al., 2004). Even when granted that the recognition
specificity of intact proteins by SH3 domains is greater than for
SH3-peptide recognition, affinity is not raised above one order of
magnitude (Arold, S. et al., 1998; Lee, C.H. et al., 1995). Moreover,
the ability of a point-mutant Src SH2 domain to effectively substitute
for the SH2 domain of the Sem-5 protein in activation of the Ras pathway
in vivo emphasises that the specificity of Sh2-mediated interactions is
not great (Marengere, L.E. et al., 1994). Consider the later statements
under the light of the fact that the affinity of the protein OppA for
its ligands is in the range of two orders of magnitude (Oppa is involved
in the mopping of peptides in the bacterial periplasm exhibiting no
sequence specificity). Tu put it all into a nutshell: the described
facts lead to a view of large and promiscuous SH-mediated interaction
networks.
Since it is possible to generate mutant SH3 domains
that have up to 40-fold higher affinity than their wild-types (Hiipakka
et al, 1999) , the potential of these domains as research tools and as
source of lead compounds for pharmaceutical development can not be
overseen. Furthermore, a question cannot be overheard in our minds:
which is the functional advantage of maintaining relative low affinity
and selectivity for PID-mediated interactions, instead of optimizing the
potential affinity of PIDs? And further: how can PID-dependent
interaction pathways achieve precise cellular responses?
A comfortable view is that sufficient effective selectivity can be brought
by compartmentalization, additive effects of multiple separate
interactions, cooperative assembly of multiprotein complexes, etc. and
that this effects can sustain linear functional pathways. An interesting
insight into this mater has been advanced by Zarrinpar, A. et al. In
their work (2003) the authors find out that while metazoan SH3 domains
may rescue the functionality of mutated Sho1-SH3 of the yeast, in the
set of yeast-own SH3 domains this promiscuity is forbidden. They thus
conclude and confirm that, on the background of diverging SH3 domains,
negative selection has drifted the ligand sequences to non-overlapping
areas of the particular SH3 binding regions on the sequence space.
Alternatively, a divergence process of the shape of the binding region
may be ‘guided’ by positive selection to avoid overlapping and, thus,
promiscuitive interactions. Nevertheless, the picture of linear functional
pathways is being revolutionized by a more probabilistic view of a dynamical
equilibrium between multiple interactions, in which “the central organizing
principle is a vast and ever-shifting web of interactions, from which output
is gauged by global changes in complex binding equilibria” (Mayer, B. J., 2001).
Founding
Who's visiting
start on 16.03.2009
<html> <a href="http://www2.clustrmaps.com/counter/maps.php?url=http://openwetware.org/wiki/Molecular_Recognition_Laboratorium" id="clustrMapsLink"><img src="http://www2.clustrmaps.com/counter/index2.php?url=http://openwetware.org/wiki/Molecular_Recognition_Laboratorium" style="border:0px;" alt="Locations of visitors to this page" title="Locations of visitors to this page" id="clustrMapsImg" onerror="this.onerror=null; this.src='http://clustrmaps.com/images/clustrmaps-back-soon.jpg'; document.getElementById('clustrMapsLink').href='http://clustrmaps.com';" /> </a> </html>
Science in the city: A small series about the everyday life of scientists in berlin
FOLLOW ME >>>>