M465:IndependentProject: Difference between revisions
No edit summary |
|||
| (14 intermediate revisions by 2 users not shown) | |||
| Line 15: | Line 15: | ||
<br> | <br> | ||
== Tuesday October 25th: Set Up Flies == | |||
Today we'll be establishing our flies in each of our conditions. Below are protocols for each of the three variables above. Remember that each team will focus on one of these variables. Each student will have their own set of three experimental lines for this experiment. <br> | Today we'll be establishing our flies in each of our conditions. Below are protocols for each of the three variables above. Remember that each team will focus on one of these variables. Each student will have their own set of three experimental lines for this experiment. <br> | ||
| Line 35: | Line 38: | ||
3. Using a plastic graduated pipette, take 5 mL of the food and place into one of the previously used glass vials. Label this vial CONTROL. Create four control vials <br> | 3. Using a plastic graduated pipette, take 5 mL of the food and place into one of the previously used glass vials. Label this vial CONTROL. Create four control vials <br> | ||
4. Calculate the amount of antibiotic needed for the remaining food. For example, if you have four people in your group, you will have liquified total of 60 mL of food, out of which you will have used 20 mL for the CONTROL vials. That would leave 40 mL. Remove 20 mL of food and place in a new beaker. <br> | 4. Calculate the amount of antibiotic needed for the remaining food. For example, if you have four people in your group, you will have liquified total of 60 mL of food, out of which you will have used 20 mL for the CONTROL vials. That would leave 40 mL. Remove 20 mL of food and place in a new beaker. <br> | ||
5. You are provided with | 5. You are provided with 15 mg/mL tetracycline stock. Create a food mixture with 5 ug/mL tetracycline and one with 50 ug/mL tetracycline. Stir well and dispense using the graduated pipettes. <br> | ||
6. Each student in the group should have one control, one 5 ug/mL vial and one 50 ug/mL vial. <br> | 6. Each student in the group should have one control, one 5 ug/mL vial and one 50 ug/mL vial. <br> | ||
| Line 48: | Line 51: | ||
3. Make sure your vials are labeled properly with your initials, your team, and the condition being tested! | 3. Make sure your vials are labeled properly with your initials, your team, and the condition being tested! | ||
==Thursday, October 27th: CFU analysis== | |||
Today you will perform your first analysis of colony forming units for your adult flies in each condition. Remember that we are going to be looking at the offspring of these flies for our experimental results. Looking at the adults now, especially in the control conditions, will help us establish a baseline for what type of microbiota we should expect from these flies. <br> | Today you will perform your first analysis of colony forming units for your adult flies in each condition. Remember that we are going to be looking at the offspring of these flies for our experimental results. Looking at the adults now, especially in the control conditions, will help us establish a baseline for what type of microbiota we should expect from these flies. <br> | ||
Work with your flies will be performed using the fly laboratory in Jordan Hall. <br> | Work with your flies will be performed using the fly laboratory in Jordan Hall. <br> | ||
'''Plating Fly Lysates''' | '''Plating Fly Lysates''' | ||
1. Obtain your fly vials from each of your three conditions. For each of these vials, you will plate one fly lysate. Ahead of time, label three 1.5 mL tubes with the condition and fill each tube with 500 ul of PBS <br> | 1. Obtain your fly vials from each of your three conditions. For each of these vials, you will plate one fly lysate. Ahead of time, label three 1.5 mL tubes with the condition and fill each tube with 500 ul of PBS <br> | ||
2. Collect three female flies from each of your vials using the CO2 pad and dissecting scope. <br> | 2. Collect three female flies from each of your vials using the CO2 pad and dissecting scope. All parental flies can be discarded at this point. <br> | ||
3. Place the three flies in the appropriate 1.5 mL tube with 500 ul of PBS. <br> | 3. Place the three flies in the appropriate 1.5 mL tube with 500 ul of PBS. <br> | ||
4. Homogenize the flies in PBS using a sterile pestle. <br> | 4. Homogenize the flies in PBS using a sterile pestle. <br> | ||
| Line 66: | Line 70: | ||
10. Uncap your agar plate. Take 200 ul of your dilutions (tube 2) and spot it onto the center of the agar. Using sterile glass beads, spread the liquid as evenly as possible across the surface of the agar. <br> | 10. Uncap your agar plate. Take 200 ul of your dilutions (tube 2) and spot it onto the center of the agar. Using sterile glass beads, spread the liquid as evenly as possible across the surface of the agar. <br> | ||
11. Replace the cap and allow your sample to sit, undisturbed, agar side down, for ~2 minutes. <br> | 11. Replace the cap and allow your sample to sit, undisturbed, agar side down, for ~2 minutes. <br> | ||
12. Repeat the plating and spreading for dilution tube 3 and for each of the two media types. | 12. Repeat the plating and spreading for dilution tube 3 and for each of the two media types. <br> | ||
You will incubate your fly lysates at 30C. <br> | |||
'''Remember to come back to the laboratory on Saturday to count colonies!''' <br> | |||
=='''Isolating DNA from your Insects'''== | |||
Last time we met you cultured microbes from your experimental flies. Today, we will start the process of using a culture independent approach to identifying the microbes in your samples. First, we will liberate the DNA in your sample, then, the following meeting, we will amplify the 16S rRNA gene from your samples. Finally, the following week, we will clone that gene into a plasmid, allowing us to sample the 16S rRNA genes in your amplicon by sequencing. <br> <br> | |||
'''Isolate Genomic DNA From Your Sample'''<BR> | |||
''Please wear gloves during this protocol''<br><br> | |||
*This protocol you've done before! Should be quick and easy! | |||
1. Last time we met you transfered 3 flies from each of your vials into a labeled, 1.5 mL tube. These were kept at -20C until today. <br> | |||
2. To each of the tubes, add 200 uL of PBS and 20 uL of proteinase K. <br> | |||
3. Using a sterile pestle, grind the fly sample in the tube as well as you can. <br> | |||
4. Add 200 uL of buffer ATL to each 1.5 mL tube and mix by vortexing. <br> | |||
5. Incubate your mixture at 56C for 10 minutes (the water bath in the back). <br> | |||
6. Once the incubation is done, add 200 ul of ethanol (100%) to the suspension and mix by vortexing. <br> | |||
7. For each tube, label an individual DNeasy mini spin column in a 2mL collection tube. <br> | |||
8. Pipet the entire mixture into the appropriately labeled column. <br> | |||
9. Spin the column at 6,000 g for 1 minute. Your DNA will now have adhered to the column. Discard the flow through and place the column in a new 2 mL collection tube. <br> | |||
*the next steps are all washes * <br> | |||
10. Add 500 ul AW1 to the column. Centrifuge for 1 minute at 6,000 g. Discard the flow through and '''place the column in a new 2 mL collection tube''' It is extremely important that you place the column into a new tube as you will carry over the washes at each step otherwise.<br> | |||
11. Add 500 ul AW2 to the column and centrifuge for 3 minutes at max speed. Discard the flow through and place the column in a labled, 1.5 mL tube <br> | |||
12. Elute the DNA from the column by adding 200 ul of buffer AE to the center of the spin column. Incubate the column for 1 minute at room temperature and spin at max speed for 1 minute. <br> | |||
'''Your DNA is now in the liquid that came through the column!''' | |||
=='''Amplifying your 16S rRNA gene'''== | |||
We will next attempt to the 16S rRNA gene from your samples. After 30 cycles of polymerase chain reaction in a thermal cycler, the result will be a pcr product containing hundreds, if not thousands, of the 16S rRNA gene, essentially a population of amplified 16S fragments, each of which could have come from a different microbial template. | |||
To review how the polymerase chain reaction works and how it exponentially amplifies specific sequences of DNA, go to the following web site:<BR> | |||
PCR animation | |||
http://www.dnalc.org/resources/animations/pcr.html<BR><BR> | |||
All PCR reactions require a thermal cycler to elevate and reduce the reaction temperature quickly and keep it at a specific temperature for a prescribed amount of time. There is a basic pattern to these temp. cycles, but there are differences, so you must be sure to program the cycler with the correct time and temperature for your specific amplification. Traditionally, pcr used Taq polymerase, a heat stable DNA polymerase originally found in a extremophilic bacterium, ''Thermus aquaticus'', that lives and reproduces in boiling hot springs. We are not using Taq for our pcr but a different polymerase, Finnzyme's Phusion High-Fidelity Polymerase, a proprietary reagent that uses a novel heat-stable ''Pyrococcus-like'' enzyme. Phusion DNA Polymerase generates long templates with a greater accuracy and speed than with Taq. The error rate of Phusion DNA Polymerase in Phusion HF Buffer is determined to be 4.4 x 10-7, which is approximately 50-fold lower than that of ''Thermus aquaticus'' DNA polymerase, and 6-fold lower than that of ''Pyrococcus furiosus'', another proof-reading DNA polymerase. | |||
Therefore, our pcr product DNA will have far fewer "mistakes" in the sequences that are replicated from template DNA. Our polymerase will also work much faster so our ~20 cycles will require less time than conventional Taq based pcr. <br><br> | |||
'''Protocol for PCR'''<BR> | |||
1. Obtain one 0.2ml pcr tube from your instructor - you will need one for each of your DNA extractions (for each of your insect "types"). All of the ingredients listed below in the table, except the template DNA, have been added together previously and kept on ice for you in these tubes. <BR><BR> | |||
2. Label the tube with a fine tipped Sharpie on the side - make sure you keep track of the code name in your lab notebook. '''Do not use tape!''' <BR><BR> | |||
3. To each tube, you will add 4 μL of the DNA you extracted. Since your pcr tube already has 10μL master mix, 4μL DNAase free water, and 1μL of each of 2 primers, the total reaction volume for everyone will be 20μL.'''<BR><BR> | |||
'''It is very important to pipet these tiny volumes accurately. Use the P10 or P20 pipettes. Look at the tip after you draw up your measured volume to make sure you have liquid there.''' <BR><BR> | |||
4. Dispense the template DNA into the liquid directly, watching to make sure that the liquid has left the pipette tip. <BR><BR> | |||
5. Bring your tube to your instructor; they will show you where the thermal cycler is located in JH 022. Keep track of where in the PCR machine your tubes have been placed (the exact quadrant, row and column). Your instructor will start the reaction when everyone's tubes are loaded. <BR><BR> | |||
'''Component TABLE '''<BR> | |||
{| border="1" | |||
|+ | |||
! Component !! amt. in a 20 μl<br>reaction !! Final Conc. | |||
|- | |||
! Purified<BR>DNAase free <BR> Water | |||
| 4 μL already in tube.<BR> Want to achieve<br>total of 20 μl reaction vol.<br> Add from 0 - 3μl | |||
| _ | |||
|- | |||
! 2x Phusion Master Mix | |||
| 10 μl | |||
| 1x | |||
|- | |||
! 27F primer | |||
| 1 | |||
| 0.5 μMolar | |||
|- | |||
! 1492R primer | |||
| 1 | |||
| 0.5 μMolar | |||
|- | |||
! template DNA | |||
| 4 μl | |||
| optimum is 100ng of DNA/reaction | |||
|- | |||
|} | |||
The cycling program is shown below. <BR><BR> | |||
94°C for 2 min | |||
37 cycles at: | |||
94°C for 30 s | |||
59°C for 45 s | |||
72°C for 1 min 30 s | |||
1 cycle at: | |||
72°C for 10 min | |||
4°C hold | |||
==Agarose Gel Electrophoresis of Clean PCR PRODUCT== | |||
To see if you successfully amplified the 16S rRNA gene and not anything else, you will "run a gel" on your cleaned pcr products. To run a gel means that we will perform an electrophoretic separation of the DNA fragments in your cleaned up pcr product, using 5uL vol. of your pcr product applied to a 1% agarose gel stained with Sybr Safe DNA stain. Your instructor will photograph the gel, label it with your amplicon id from the template and post the gel photo on Canvas so you can evaluate your success at 16S rRNA gene amplification. You should see a single band of ~1.5kb indicating that the only dsDNA in your pcr product came from amplification of a ~1500bp gene fragment. Can you explain how we know the size of our amplified gene fragment?<BR><BR> | |||
Your agarose gel is made of 1.0% agarose (w/v) in 1x TBE buffer (10x=890mM Tris, 890mM Boric Acid, 20mM EDTA) with SybrSafe™ stain.<BR><BR> | |||
DNA is uniformly negatively charged and will,therefore, move toward the positive electrode. The separation is determined by the size or mass of the molecule or fragments of DNA. <BR><BR> | |||
[[Image:BISC110_gel2.jpg]]<BR><BR> | |||
''' Procedure for Agarose Gel Electrophoresis of PCR products'''<BR> | |||
You will put the 5 microliters of your pcr product as a spot on a small piece of parafilm and add 5 microliters of loading dye (0.25% XC, 30% glycerol, 0.1mg/ml RNAase). '''Mix the loading dye''' by pipetting up and down before loading all 10 microliters into a lane of the 1% agarose gel (1% wt/vol in 1xTBE buffer with Sybr Safe DNA stain (a proprietary reagent from Invitrogen used according to manufacturer's directions at http://www.invitrogen.com). Record on the gel template in which well you have loaded your pcr product. Be sure to leave the first lane empty for the 1kbp ladder. <BR><BR> | |||
Note that Loading dye contains glycerol to keep our sample in the lane rather than floating away and will have one of 3 marker dyes (bromophenol blue, xylene cyanol, or orange G) that facilitate estimation of DNA migration distance and optimization of agarose gel run time. 1x TBE buffer is used in this electrophoretic separation (89mM Tris, 89mM Boric acid, 2.0mM EDTA. The gel will be run at 120V for approximately 30 minutes. <BR><BR> | |||
How will you judge a successful amplification? How many fragments and of what size do you expect to see? <BR><BR> | |||
=='''Clean up of your PCR product'''== | |||
Before we can sesquence our bacterial 16S rRNA genes, we must remove interfering dNPTs, primers, and other small degraded DNA. We will use a column that separates DNA by size. Since the reagents and column materials in the kit we will use are proprietary, we won't know exactly what is going on at each step but, basicially, we will apply our pcr product to a column of a particular density, wash away elements too small to be trapped in it, and elute off the larger fragments of DNA (that should be ~1500bps if our pcr amplification of the 16s rRNA genes in our soil genomic DNA was successful). | |||
'''Notes before Starting:'''<BR> | |||
95% ethanol has been added to Buffer PE before first time use (see bottle label for volume).<BR> | |||
All centrifuge steps are carried out at 17,900rfc (~13,000 rpm in a microcentrifuge) in a conventional tabletop microcentrifuge at room temperature.<BR> | |||
You will be performing the procedure below for each PCR reaction you performed - for five isolates, that is 5 total! <br> | |||
'''Procedure'''<BR> | |||
1. Add 5 volumes of Buffer PB to 1 volume of the PCR reaction and mix. Because our PCR reactions are 20 ul volume, we will be adding 100 ul of buffer PB to the tubes. <br> <br> | |||
2. Place a QIAquick column in a 2 ml collection tube. <br> <br> | |||
3. To bind the DNA to the column, apply the sample to the surface of the QIAquick column and centrifuge for 30-60 seconds. <br> <br> | |||
4. Discard the flow-through and place the QIAquick column back in the same tube. <br> <br> | |||
5. To wash, add 750 ul of buffer PE to the QIAquick column and centrifuge for 30-60 s. Discard the flow-through and place the column back in the same tube. <br> <br> | |||
6. Centrifuge the column once more for 30-60s in the provided 2 ml tube to remove residual wash buffer. <br> <br> | |||
7. Place each QIAquick column in a clean, 1.5 mL microcentrifuge tube. <br> <br> | |||
8. To elute DNA, add 50 ul of buffer EB to the surface of the column - make sure to place this volume at the center of the membrane. Centrifuge the column for 1 minute. <br> <br> | |||
9. The purified DNA should now be at the bottom of your 1.5 mL tube. Discard the column. <br> <br> | |||
''IMPORTANT NOTES for using this kit: Ensure that the elution buffer (EB) is dispensed directly onto the spin column membrane for complete elution of bound DNA. The average eluate volume is 48 μl from 50 μl elution buffer volume.''<BR> | |||
''Elution efficiency is dependent on pH. The maximum elution efficiency is achieved | |||
between pH 7.0 and 8.5. Store DNA at –20°C as DNA may degrade in the absence of a buffering | |||
agent. ''<BR><BR> | |||
Make sure your pcr product is clearly labeled! | |||
==Culture-Independent Identification of Soil Bacteria== | |||
Now that we have cleaned our pcr products containing amplified fragments of 16s rRNA gene from many of the species of bacteria in your fly sample, today you will insert your bacterial 16s rRNA gene fragments into a patented cloning vector (pCR-BluntII TOPO®) and then transform that vector into a special genetically engineered strain of ''Escherichia coli'' bacteria that will express a vector gene for kanamycin resistance, allowing us to select for transformants on media containing kanamycin. | |||
<BR><BR> | |||
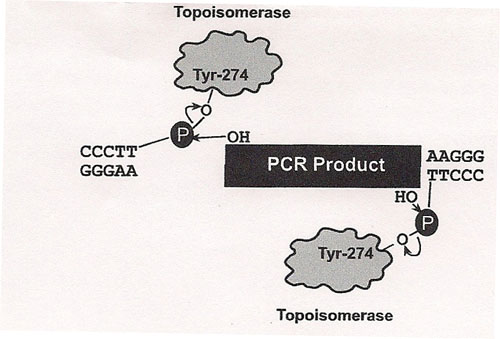
The principle behind TOPO® cloning is the enzyme DNA topoisomerase I, which will function in this system both as a restriction enzyme and as a ligase. Its biological role is to cleave and rejoin DNA during replication. Vaccinia virus topoisomerase I specifically recognizes the pentameric sequence 5´-(C/T)CCTT-3´ and forms a covalent bond with the | |||
phosphate group attached to the 3´ thymidine. It cleaves one DNA strand, enabling the DNA to unwind. The enzyme then religates the ends of the cleaved strand and releases itself from the DNA. To harness the religating activity of topoisomerase, TOPO® vectors are provided linearized with topoisomerase I covalently bound to each 3´ phosphate. This enables the vectors to quickly ligate DNA sequences with compatible ends. <BR><BR> | |||
[[Image:toposchemablunt.jpg]] | |||
We used a polymerase that creates blunt ended DNA fragments rather than using TaQ. Taq polymerase makes fragments with 3' T overhangs; therefore, complementary single stranded A rich "sticky ends" allow ligation. Blunt ends require a different Blunt-fragment cloning protocol. Invitrogen's Zero Blunt® TOPO® PCR Cloning Kit will work well for us. It has several (T7, SP6, and M13 forward and reverse) priming sites for directing sequencing to the appropriate region and it has two resistance genes, Kanamycin and Zeocin, for selecting clones in a genetically engineered form of ''E. coli'' that we will use for separating the amplified 16s rRNA genes from our soil flora.<BR><BR> | |||
[[Image:pcr_Blunt.jpg]] | |||
Additionally, the cloning system we will use contains two different background reducers, one of which is a lethal ccdB (control of cell death)gene encoding a ccdB protein that poisons bacterial DNA gyrase, causing degradation of the host chromosome and cell death. When one of our 16s rRNA genes from our pcr product is ligated into the vector, the ccdB gene is disrupted, enabling recombinant colonies to grow while other colonies without a vector insert will not grow. Because a few colonies may form despite the undisrupted expression of ccdB there is a second mechanism of insuring that we only pick colonies coming from cells with our 16s rRNA gene insert. | |||
==Using Zero Blunt TOPO PCR Cloning Kit with One Shot TOP 10 Chemically Competent ''E. coli''== | |||
PCR cloning requires three steps. <br> | |||
[[Image:protocloning2.jpg]] | |||
<BR><BR> | |||
We will clone your three pcr products individually, if you had three successful amplifications. If you had 2 successful amplifications from your sampling site, use only the most successful 16s rRNA gene amplifications and omit the one that did not work. If you did not get amplification, you will repeat your PCR amplification today.<BR><BR> | |||
Procedure: Your instructor will have added the following reagents in this order to a 0.2 mL tube for you<BR> | |||
1. Add 0.5 μL of salt solution (final conc. 200mM NaCl, 10mM MgCl<sub>2</sub>).<BR> | |||
2. Add 1 μL of purified water (DNAase free).<BR> | |||
3. Add 0.5 μL of pCR®II-Blunt-TOPO® cloning vector plasmid. (MAKE sure you pipet this correctly with a P2 and a filter tip!)<BR> | |||
Your job is to add 1 uL of your cleaned PCR products to the above reaction mix. You will need three mixes for a total of three cloning reactions (one for each fly experimental condition). After you have added your PCR products, incubate the reaction for 15 min at room temperature.<BR> | |||
Continue to next step: Transform Oneshot Top10 competent ''E. coli''.<br><BR> | |||
==Transforming TOPO Competent ''E. coli''== | |||
'''Genotype of OneShot TOP10 Competent Cells:''' ''F- mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(araleu) 7697 galU galK rpsL (StrR) endA1 nupG''<BR><BR> | |||
'''General Handling''': Be extremely gentle when working with competent cells. Competent cells have been chemically treated to make their cell walls and membranes more porous so they are fragile and highly sensitive to changes in temperature. They can be easily lysed by too vigorous pipetting. Transformation should be started immediately following the thawing of the cells on ice. Mix by swirling or tapping the tube gently, not by pipetting(no vortexing). <BR><BR> | |||
'''Transforming One Shot® Competent Cells'''<BR> | |||
'''Introduction''': Once you have performed the TOPO® Cloning reaction, you will transform your pCR®-Blunt II-TOPO® construct into TOPO10 competent E. coli provided with your kit.<BR> | |||
'''You will need the following reagents and equipment:'''<BR> | |||
• TOPO® Cloning reaction from Performing the TOPO® Cloning Reaction<BR> | |||
• S.O.C. medium (super optimal broth medium:0.5% Yeast Extract;2% Tryptone;10 mM NaCl;2.5 mM KCl;10 mM MgCl2;10 mM MgSO4;20 mM Glucose)This medium is included with the kit)<BR> | |||
• 42°C water bath or heat block<BR> | |||
• WARM Luria-Bertoni (LB) solid medium containing 50 μg/ml kanamycin <BR> | |||
• 37°C shaking and non-shaking incubators<BR><BR> | |||
**every student will need an ice bucket!** <br> | |||
'''Preparing for Transformation'''<BR> | |||
For each transformation, you will need one vial of competent cells and two | |||
selective medium agar plates.<BR> | |||
• Equilibrate a water bath to 42°C<BR> | |||
• Bring the vial of S.O.C. medium to room temperature.<BR> | |||
• Warm LB plates containing 50 μg/ml kanamycin at 37°C | |||
for 30 minutes.<BR> | |||
• Thaw on ice 1 vial of One Shot® cells for each transformation on ice. These will be ready for you at the front of the room. <BR><BR> | |||
'''Transformation Procedure'''<BR> | |||
1. Add 2 μl of the TOPO® Cloning reaction when it is completed into a vial of One Shot® Chemically Competent ''E. coli'' and mix gently by swirling. '''Do not mix by pipetting up and down!'''<BR><BR> | |||
2. Incubate on ice for 10 minutes. <BR><BR> | |||
3. Heat-shock the cells for 30 seconds exactly at 42°C in the water bath (without shaking).<BR><BR> | |||
4. Immediately (take your ice bucket with you to the heat block) transfer the tubes to ice .<BR><BR> | |||
5. Add 250 μl of room temperature S.O.C. medium (it must NOT be cold).<BR><BR> | |||
6. Cap the tube tightly and put the capped tube in a empty non-sterile 15 ml. conical tube and shake the tube horizontally (200 rpm) at 37°C for | |||
1 hour. | |||
7. After the 1 hour incubation of the transformation mix, Use your P200 micropipet to pipet 50 μl from each transformation to the center of a ''prewarmed'' LB + kan plate. Using glass beads, carefully spread the aliquot of cells over the entire surface of the plate.<BR><BR> | |||
8. Repeat step 7 on a new LB + kan plate, using a 200 μL volume of transformed cells. You will plate two different volumes to ensure that at least one plate will have well-spaced colonies.<BR><BR> | |||
9. Incubate all plates upside down overnight at 37°C. Remember to label each plate with all the appropriate information: your initials, your team name, the sample id, and the volume plated. | |||
10. Check your transformations after 12-18 hours (overnight incubation)to be sure of successful transformation. When medium size, ISOLATED colonies, have appeared, refrigerate your transformation plates in parafilm, in the deli case in the back of the room, until the next lab. DO NOT LEAVE THEM INCUBATING TOO LONG, resulting in overgrown colonies that are not isolated! | |||
Latest revision as of 06:46, 3 November 2016
The Effect of Environment on the Microbiota
For your independent project, you have chosen to fund a proposal focused on environmental variables that may alter or impact the Drosophila microbiota. Each team will focus on one of the three variables below:
1) Temperature (18C, 25C, 28C)
2) Glyphosate (0 ug/mL, 0.1 ug/mL, 0.5 ug/mL)
3) Antibiotics (0 ug/mL, 5 ug/mL, 50 ug/mL)
For each variable, you will have three conditions (outlined above in parentheses). You will begin with wild type flies lacking any Wolbachia infection.
Below is a rough outline of the next few weeks - things may be adjusted depending on how well the flies grow, etc.
Tuesday October 25th: Set Up Flies
Today we'll be establishing our flies in each of our conditions. Below are protocols for each of the three variables above. Remember that each team will focus on one of these variables. Each student will have their own set of three experimental lines for this experiment.
Glyphosate
- Protocol to be performed as a group such that you standardize your food across the team.
1. Each student will collect three vials of food from your instructor. These vials have 5 mL of food in them each. In addition, you will need two small beakers.
2. Microwave the food vials for 10 seconds at a time until the food is liquified. Pour the food into the small beaker.
3. Using a plastic graduated pipette, take 5 mL of the food and place into one of the previously used glass vials. Label this vial CONTROL. Create four control vials
4. Calculate the amount of glyphosate needed for the remaining food. For example, if you have four people in your group, you will have liquified total of 60 mL of food, out of which you will have used 20 mL for the CONTROL vials. That would leave 40 mL. Remove 20 mL of food and place in a new beaker.
5. Create a food mixture with 0.1 ug/mL glyphosate and one with 0.5 ug/mL glyphosate. Stir well and dispense using the graduated pipettes.
6. Each student in the group should have one control, one 0.1 ug/mL vial and one 0.5 ug/mL vial.
Antibiotic treatment
- Protocol to be performed as a group such that you standardize your food across the team.
1. Each student will collect three vials of food from your instructor. These vials have 5 mL of food in them each. In addition, you will need two small beakers.
2. Microwave the food vials for 10 seconds at a time until the food is liquified. Pour the food into the small beaker.
3. Using a plastic graduated pipette, take 5 mL of the food and place into one of the previously used glass vials. Label this vial CONTROL. Create four control vials
4. Calculate the amount of antibiotic needed for the remaining food. For example, if you have four people in your group, you will have liquified total of 60 mL of food, out of which you will have used 20 mL for the CONTROL vials. That would leave 40 mL. Remove 20 mL of food and place in a new beaker.
5. You are provided with 15 mg/mL tetracycline stock. Create a food mixture with 5 ug/mL tetracycline and one with 50 ug/mL tetracycline. Stir well and dispense using the graduated pipettes.
6. Each student in the group should have one control, one 5 ug/mL vial and one 50 ug/mL vial.
Temperature
1. Each student in the group should collect three vials of food from your instructor.
2. Label each vial with the temperature: either 18C, 25C or 28C.
Collecting your flies
- This part of the protocol will be performed upstairs in the Newton Laboratory after your vials ready.
1. For each of your vials you will need 10 female flies and 5 male flies. Collect these on the CO2 pad and place them in your vials to wake.
2. After you have established your lines, place them in the Newton Lab incubator (for tetracycline and glyphosate treatments) or in the temperature incubators appropriate to the treatment (for the temperature experiment).
3. Make sure your vials are labeled properly with your initials, your team, and the condition being tested!
Thursday, October 27th: CFU analysis
Today you will perform your first analysis of colony forming units for your adult flies in each condition. Remember that we are going to be looking at the offspring of these flies for our experimental results. Looking at the adults now, especially in the control conditions, will help us establish a baseline for what type of microbiota we should expect from these flies.
Work with your flies will be performed using the fly laboratory in Jordan Hall.
Plating Fly Lysates
1. Obtain your fly vials from each of your three conditions. For each of these vials, you will plate one fly lysate. Ahead of time, label three 1.5 mL tubes with the condition and fill each tube with 500 ul of PBS
2. Collect three female flies from each of your vials using the CO2 pad and dissecting scope. All parental flies can be discarded at this point.
3. Place the three flies in the appropriate 1.5 mL tube with 500 ul of PBS.
4. Homogenize the flies in PBS using a sterile pestle.
5. For each condition, perform a dilution series. Label three tubes 1,2,3. Add 0.9 mL of PBS to each of these tubes.
6. Add 0.1 mL of your original fly lysate to tube 1. Cap the tube and mix by vortexing.
7. Add 0.1 mL of the liquid in tube 1 to the liquid in tube 2. Cap tube 2 and mix by vortexing. Tube 2 will be our 10^2 dilution.
8. Add 0.1 mL of the liquid in tube 2 to the liquid in tube 3. Cap tube 3 and mix by vortexing. Tube 3 will be our 10^3 dilution.
9. At this point you are ready to plate your dilutions. For each condition, label four solid agar plates (two of MRS and two of LB, you will plate on both) with your initials, team name, the experimental condition, and the dilution factor. "make sure you label the agar side!". You should have a total of 12 plates at this point.
10. Uncap your agar plate. Take 200 ul of your dilutions (tube 2) and spot it onto the center of the agar. Using sterile glass beads, spread the liquid as evenly as possible across the surface of the agar.
11. Replace the cap and allow your sample to sit, undisturbed, agar side down, for ~2 minutes.
12. Repeat the plating and spreading for dilution tube 3 and for each of the two media types.
You will incubate your fly lysates at 30C.
Remember to come back to the laboratory on Saturday to count colonies!
Isolating DNA from your Insects
Last time we met you cultured microbes from your experimental flies. Today, we will start the process of using a culture independent approach to identifying the microbes in your samples. First, we will liberate the DNA in your sample, then, the following meeting, we will amplify the 16S rRNA gene from your samples. Finally, the following week, we will clone that gene into a plasmid, allowing us to sample the 16S rRNA genes in your amplicon by sequencing.
Isolate Genomic DNA From Your Sample
Please wear gloves during this protocol
- This protocol you've done before! Should be quick and easy!
1. Last time we met you transfered 3 flies from each of your vials into a labeled, 1.5 mL tube. These were kept at -20C until today.
2. To each of the tubes, add 200 uL of PBS and 20 uL of proteinase K.
3. Using a sterile pestle, grind the fly sample in the tube as well as you can.
4. Add 200 uL of buffer ATL to each 1.5 mL tube and mix by vortexing.
5. Incubate your mixture at 56C for 10 minutes (the water bath in the back).
6. Once the incubation is done, add 200 ul of ethanol (100%) to the suspension and mix by vortexing.
7. For each tube, label an individual DNeasy mini spin column in a 2mL collection tube.
8. Pipet the entire mixture into the appropriately labeled column.
9. Spin the column at 6,000 g for 1 minute. Your DNA will now have adhered to the column. Discard the flow through and place the column in a new 2 mL collection tube.
- the next steps are all washes *
10. Add 500 ul AW1 to the column. Centrifuge for 1 minute at 6,000 g. Discard the flow through and place the column in a new 2 mL collection tube It is extremely important that you place the column into a new tube as you will carry over the washes at each step otherwise.
11. Add 500 ul AW2 to the column and centrifuge for 3 minutes at max speed. Discard the flow through and place the column in a labled, 1.5 mL tube
12. Elute the DNA from the column by adding 200 ul of buffer AE to the center of the spin column. Incubate the column for 1 minute at room temperature and spin at max speed for 1 minute.
Your DNA is now in the liquid that came through the column!
Amplifying your 16S rRNA gene
We will next attempt to the 16S rRNA gene from your samples. After 30 cycles of polymerase chain reaction in a thermal cycler, the result will be a pcr product containing hundreds, if not thousands, of the 16S rRNA gene, essentially a population of amplified 16S fragments, each of which could have come from a different microbial template.
To review how the polymerase chain reaction works and how it exponentially amplifies specific sequences of DNA, go to the following web site:
PCR animation
http://www.dnalc.org/resources/animations/pcr.html
All PCR reactions require a thermal cycler to elevate and reduce the reaction temperature quickly and keep it at a specific temperature for a prescribed amount of time. There is a basic pattern to these temp. cycles, but there are differences, so you must be sure to program the cycler with the correct time and temperature for your specific amplification. Traditionally, pcr used Taq polymerase, a heat stable DNA polymerase originally found in a extremophilic bacterium, Thermus aquaticus, that lives and reproduces in boiling hot springs. We are not using Taq for our pcr but a different polymerase, Finnzyme's Phusion High-Fidelity Polymerase, a proprietary reagent that uses a novel heat-stable Pyrococcus-like enzyme. Phusion DNA Polymerase generates long templates with a greater accuracy and speed than with Taq. The error rate of Phusion DNA Polymerase in Phusion HF Buffer is determined to be 4.4 x 10-7, which is approximately 50-fold lower than that of Thermus aquaticus DNA polymerase, and 6-fold lower than that of Pyrococcus furiosus, another proof-reading DNA polymerase.
Therefore, our pcr product DNA will have far fewer "mistakes" in the sequences that are replicated from template DNA. Our polymerase will also work much faster so our ~20 cycles will require less time than conventional Taq based pcr.
Protocol for PCR
1. Obtain one 0.2ml pcr tube from your instructor - you will need one for each of your DNA extractions (for each of your insect "types"). All of the ingredients listed below in the table, except the template DNA, have been added together previously and kept on ice for you in these tubes.
2. Label the tube with a fine tipped Sharpie on the side - make sure you keep track of the code name in your lab notebook. Do not use tape!
3. To each tube, you will add 4 μL of the DNA you extracted. Since your pcr tube already has 10μL master mix, 4μL DNAase free water, and 1μL of each of 2 primers, the total reaction volume for everyone will be 20μL.
It is very important to pipet these tiny volumes accurately. Use the P10 or P20 pipettes. Look at the tip after you draw up your measured volume to make sure you have liquid there.
4. Dispense the template DNA into the liquid directly, watching to make sure that the liquid has left the pipette tip.
5. Bring your tube to your instructor; they will show you where the thermal cycler is located in JH 022. Keep track of where in the PCR machine your tubes have been placed (the exact quadrant, row and column). Your instructor will start the reaction when everyone's tubes are loaded.
Component TABLE
| Component | amt. in a 20 μl reaction |
Final Conc. |
|---|---|---|
| Purified DNAase free Water |
4 μL already in tube. Want to achieve total of 20 μl reaction vol. Add from 0 - 3μl |
_ |
| 2x Phusion Master Mix | 10 μl | 1x |
| 27F primer | 1 | 0.5 μMolar |
| 1492R primer | 1 | 0.5 μMolar |
| template DNA | 4 μl | optimum is 100ng of DNA/reaction |
The cycling program is shown below.
94°C for 2 min
37 cycles at:
94°C for 30 s
59°C for 45 s
72°C for 1 min 30 s
1 cycle at:
72°C for 10 min
4°C hold
Agarose Gel Electrophoresis of Clean PCR PRODUCT
To see if you successfully amplified the 16S rRNA gene and not anything else, you will "run a gel" on your cleaned pcr products. To run a gel means that we will perform an electrophoretic separation of the DNA fragments in your cleaned up pcr product, using 5uL vol. of your pcr product applied to a 1% agarose gel stained with Sybr Safe DNA stain. Your instructor will photograph the gel, label it with your amplicon id from the template and post the gel photo on Canvas so you can evaluate your success at 16S rRNA gene amplification. You should see a single band of ~1.5kb indicating that the only dsDNA in your pcr product came from amplification of a ~1500bp gene fragment. Can you explain how we know the size of our amplified gene fragment?
Your agarose gel is made of 1.0% agarose (w/v) in 1x TBE buffer (10x=890mM Tris, 890mM Boric Acid, 20mM EDTA) with SybrSafe™ stain.
DNA is uniformly negatively charged and will,therefore, move toward the positive electrode. The separation is determined by the size or mass of the molecule or fragments of DNA.

Procedure for Agarose Gel Electrophoresis of PCR products
You will put the 5 microliters of your pcr product as a spot on a small piece of parafilm and add 5 microliters of loading dye (0.25% XC, 30% glycerol, 0.1mg/ml RNAase). Mix the loading dye by pipetting up and down before loading all 10 microliters into a lane of the 1% agarose gel (1% wt/vol in 1xTBE buffer with Sybr Safe DNA stain (a proprietary reagent from Invitrogen used according to manufacturer's directions at http://www.invitrogen.com). Record on the gel template in which well you have loaded your pcr product. Be sure to leave the first lane empty for the 1kbp ladder.
Note that Loading dye contains glycerol to keep our sample in the lane rather than floating away and will have one of 3 marker dyes (bromophenol blue, xylene cyanol, or orange G) that facilitate estimation of DNA migration distance and optimization of agarose gel run time. 1x TBE buffer is used in this electrophoretic separation (89mM Tris, 89mM Boric acid, 2.0mM EDTA. The gel will be run at 120V for approximately 30 minutes.
How will you judge a successful amplification? How many fragments and of what size do you expect to see?
Clean up of your PCR product
Before we can sesquence our bacterial 16S rRNA genes, we must remove interfering dNPTs, primers, and other small degraded DNA. We will use a column that separates DNA by size. Since the reagents and column materials in the kit we will use are proprietary, we won't know exactly what is going on at each step but, basicially, we will apply our pcr product to a column of a particular density, wash away elements too small to be trapped in it, and elute off the larger fragments of DNA (that should be ~1500bps if our pcr amplification of the 16s rRNA genes in our soil genomic DNA was successful).
Notes before Starting:
95% ethanol has been added to Buffer PE before first time use (see bottle label for volume).
All centrifuge steps are carried out at 17,900rfc (~13,000 rpm in a microcentrifuge) in a conventional tabletop microcentrifuge at room temperature.
You will be performing the procedure below for each PCR reaction you performed - for five isolates, that is 5 total!
Procedure
1. Add 5 volumes of Buffer PB to 1 volume of the PCR reaction and mix. Because our PCR reactions are 20 ul volume, we will be adding 100 ul of buffer PB to the tubes.
2. Place a QIAquick column in a 2 ml collection tube.
3. To bind the DNA to the column, apply the sample to the surface of the QIAquick column and centrifuge for 30-60 seconds.
4. Discard the flow-through and place the QIAquick column back in the same tube.
5. To wash, add 750 ul of buffer PE to the QIAquick column and centrifuge for 30-60 s. Discard the flow-through and place the column back in the same tube.
6. Centrifuge the column once more for 30-60s in the provided 2 ml tube to remove residual wash buffer.
7. Place each QIAquick column in a clean, 1.5 mL microcentrifuge tube.
8. To elute DNA, add 50 ul of buffer EB to the surface of the column - make sure to place this volume at the center of the membrane. Centrifuge the column for 1 minute.
9. The purified DNA should now be at the bottom of your 1.5 mL tube. Discard the column.
IMPORTANT NOTES for using this kit: Ensure that the elution buffer (EB) is dispensed directly onto the spin column membrane for complete elution of bound DNA. The average eluate volume is 48 μl from 50 μl elution buffer volume.
Elution efficiency is dependent on pH. The maximum elution efficiency is achieved
between pH 7.0 and 8.5. Store DNA at –20°C as DNA may degrade in the absence of a buffering
agent.
Make sure your pcr product is clearly labeled!
Culture-Independent Identification of Soil Bacteria
Now that we have cleaned our pcr products containing amplified fragments of 16s rRNA gene from many of the species of bacteria in your fly sample, today you will insert your bacterial 16s rRNA gene fragments into a patented cloning vector (pCR-BluntII TOPO®) and then transform that vector into a special genetically engineered strain of Escherichia coli bacteria that will express a vector gene for kanamycin resistance, allowing us to select for transformants on media containing kanamycin.
The principle behind TOPO® cloning is the enzyme DNA topoisomerase I, which will function in this system both as a restriction enzyme and as a ligase. Its biological role is to cleave and rejoin DNA during replication. Vaccinia virus topoisomerase I specifically recognizes the pentameric sequence 5´-(C/T)CCTT-3´ and forms a covalent bond with the
phosphate group attached to the 3´ thymidine. It cleaves one DNA strand, enabling the DNA to unwind. The enzyme then religates the ends of the cleaved strand and releases itself from the DNA. To harness the religating activity of topoisomerase, TOPO® vectors are provided linearized with topoisomerase I covalently bound to each 3´ phosphate. This enables the vectors to quickly ligate DNA sequences with compatible ends.
We used a polymerase that creates blunt ended DNA fragments rather than using TaQ. Taq polymerase makes fragments with 3' T overhangs; therefore, complementary single stranded A rich "sticky ends" allow ligation. Blunt ends require a different Blunt-fragment cloning protocol. Invitrogen's Zero Blunt® TOPO® PCR Cloning Kit will work well for us. It has several (T7, SP6, and M13 forward and reverse) priming sites for directing sequencing to the appropriate region and it has two resistance genes, Kanamycin and Zeocin, for selecting clones in a genetically engineered form of E. coli that we will use for separating the amplified 16s rRNA genes from our soil flora.
Additionally, the cloning system we will use contains two different background reducers, one of which is a lethal ccdB (control of cell death)gene encoding a ccdB protein that poisons bacterial DNA gyrase, causing degradation of the host chromosome and cell death. When one of our 16s rRNA genes from our pcr product is ligated into the vector, the ccdB gene is disrupted, enabling recombinant colonies to grow while other colonies without a vector insert will not grow. Because a few colonies may form despite the undisrupted expression of ccdB there is a second mechanism of insuring that we only pick colonies coming from cells with our 16s rRNA gene insert.
Using Zero Blunt TOPO PCR Cloning Kit with One Shot TOP 10 Chemically Competent E. coli
PCR cloning requires three steps.

We will clone your three pcr products individually, if you had three successful amplifications. If you had 2 successful amplifications from your sampling site, use only the most successful 16s rRNA gene amplifications and omit the one that did not work. If you did not get amplification, you will repeat your PCR amplification today.
Procedure: Your instructor will have added the following reagents in this order to a 0.2 mL tube for you
1. Add 0.5 μL of salt solution (final conc. 200mM NaCl, 10mM MgCl2).
2. Add 1 μL of purified water (DNAase free).
3. Add 0.5 μL of pCR®II-Blunt-TOPO® cloning vector plasmid. (MAKE sure you pipet this correctly with a P2 and a filter tip!)
Your job is to add 1 uL of your cleaned PCR products to the above reaction mix. You will need three mixes for a total of three cloning reactions (one for each fly experimental condition). After you have added your PCR products, incubate the reaction for 15 min at room temperature.
Continue to next step: Transform Oneshot Top10 competent E. coli.
Transforming TOPO Competent E. coli
Genotype of OneShot TOP10 Competent Cells: F- mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(araleu) 7697 galU galK rpsL (StrR) endA1 nupG
General Handling: Be extremely gentle when working with competent cells. Competent cells have been chemically treated to make their cell walls and membranes more porous so they are fragile and highly sensitive to changes in temperature. They can be easily lysed by too vigorous pipetting. Transformation should be started immediately following the thawing of the cells on ice. Mix by swirling or tapping the tube gently, not by pipetting(no vortexing).
Transforming One Shot® Competent Cells
Introduction: Once you have performed the TOPO® Cloning reaction, you will transform your pCR®-Blunt II-TOPO® construct into TOPO10 competent E. coli provided with your kit.
You will need the following reagents and equipment:
• TOPO® Cloning reaction from Performing the TOPO® Cloning Reaction
• S.O.C. medium (super optimal broth medium:0.5% Yeast Extract;2% Tryptone;10 mM NaCl;2.5 mM KCl;10 mM MgCl2;10 mM MgSO4;20 mM Glucose)This medium is included with the kit)
• 42°C water bath or heat block
• WARM Luria-Bertoni (LB) solid medium containing 50 μg/ml kanamycin
• 37°C shaking and non-shaking incubators
- every student will need an ice bucket!**
- every student will need an ice bucket!**
Preparing for Transformation
For each transformation, you will need one vial of competent cells and two
selective medium agar plates.
• Equilibrate a water bath to 42°C
• Bring the vial of S.O.C. medium to room temperature.
• Warm LB plates containing 50 μg/ml kanamycin at 37°C
for 30 minutes.
• Thaw on ice 1 vial of One Shot® cells for each transformation on ice. These will be ready for you at the front of the room.
Transformation Procedure
1. Add 2 μl of the TOPO® Cloning reaction when it is completed into a vial of One Shot® Chemically Competent E. coli and mix gently by swirling. Do not mix by pipetting up and down!
2. Incubate on ice for 10 minutes.
3. Heat-shock the cells for 30 seconds exactly at 42°C in the water bath (without shaking).
4. Immediately (take your ice bucket with you to the heat block) transfer the tubes to ice .
5. Add 250 μl of room temperature S.O.C. medium (it must NOT be cold).
6. Cap the tube tightly and put the capped tube in a empty non-sterile 15 ml. conical tube and shake the tube horizontally (200 rpm) at 37°C for
1 hour.
7. After the 1 hour incubation of the transformation mix, Use your P200 micropipet to pipet 50 μl from each transformation to the center of a prewarmed LB + kan plate. Using glass beads, carefully spread the aliquot of cells over the entire surface of the plate.
8. Repeat step 7 on a new LB + kan plate, using a 200 μL volume of transformed cells. You will plate two different volumes to ensure that at least one plate will have well-spaced colonies.
9. Incubate all plates upside down overnight at 37°C. Remember to label each plate with all the appropriate information: your initials, your team name, the sample id, and the volume plated.
10. Check your transformations after 12-18 hours (overnight incubation)to be sure of successful transformation. When medium size, ISOLATED colonies, have appeared, refrigerate your transformation plates in parafilm, in the deli case in the back of the room, until the next lab. DO NOT LEAVE THEM INCUBATING TOO LONG, resulting in overgrown colonies that are not isolated!