M465:Communityanalyses: Difference between revisions
No edit summary |
No edit summary |
||
| (4 intermediate revisions by 2 users not shown) | |||
| Line 4: | Line 4: | ||
<div style="padding: 10px; width: 720px; border: 5px solid #B3CD4E;"> | <div style="padding: 10px; width: 720px; border: 5px solid #B3CD4E;"> | ||
==Interaction | |||
==Antibiotic | ==Antagonistic and Mutualistic Interactions== | ||
<font color = "red"> *NOTE: You must remember to set up fresh nutrient broth cultures for your isolates 1-3 days before lab to do this test!</font color = "red"> <br> | |||
The microbial community living in your environments is a complex one with many different microorganisms. As is true of any environment, these microbes interact with each other - both functionally and physically. Do selected bacteria from your community help each other or harm each other while trying to find a niche in your community? Today, you will try to answer that question by testing your cultured isolates for examples of mutualism or antagonism (co-operation or competition)by culturing them in controlled communities. Some of these bacteria may prevent the growth of others through the production of chemical inhibitors; others might promote the growth of their neighbors by producing metabolites that are needed. We are going to look for both positive and negative interactions.<br> | |||
'''PREPARING THE ISOLATES''':<BR> | |||
You will inoculate 50 µl of log phase (young culture) isolate grown in fresh nutrient broth into the assigned well(s). Once again try to control for similar numbers of organisms in your inoculum by starting with normalized optical density measurements (start with a concentration of 0.5 OD600) | |||
<BR> | |||
<font size="+1">Interaction Assay Set Up</font size="+1"> | |||
[[Image:Interactions1.jpg]] <BR> | |||
[[Image:Interactions_slide2.jpg]] <BR> | |||
<BR> | |||
You will use 64 of the wells on a 96 well plate for this assay. Each pair will use 8 unique isolates to test for interactions. Use Excel to keep track of which organisms that will be inoculated into each well as described and illustrated below.<BR> | |||
<font color = "red"> FOLLOW THE TEMPLATE CAREFULLY!!!!!! It is easy to get this inoculation messed up, but don't! </font color = "red"> | |||
<BR> | |||
[[Image:Interactions_slide3.jpg]]<BR> | |||
Transfer 50 μL of each of 8 unique isolates to be tested into the illustrated row of wells (A1 is Isolate 1, A2 is Isolate #2 etc through A8) | |||
[[Image:Interactions_slide4.jpg]] <BR> | |||
Beginning with Isolate #2, inoculate a second 50 μl of each of your isolates into the column wells B1, C1, etc. (indicated by the green color). | |||
<BR> | |||
Add 100 μL of nutrient broth to each of the wells containing your isolates (row wells A1-A8 and column wells B1-H1) | |||
<BR> | |||
Gently move the 96 well plate in a circular motion to mix.<BR> | |||
[[Image:Interactions_slide5.jpg]] <BR> | |||
<BR> | |||
Transfer 10 μl of the contents of the 7 wells - A2 (containing isolate #2 etc) through A8 to the empty wells in each column as indicated by the yellow arrows. You will need to remove the first tip from a multichannel pipette. If you are using the multichannel pipette, be sure that you work slowly and check that each pipette tip is evenly filled. You may need to tighten the tips by hand, if so be sure to only touch the part of the tip that sits on the multichannel pipette, you wouldn't want to contaminate your wells with human organsisms!<BR> | |||
<BR> | |||
[[Image:Interactions_slide6.jpg]] <BR> | |||
<BR> | |||
Transfer 10 μl of the contents of wells A1 (containing isolate #1 etc) through H1 to each well in the row as indicated by the red arrows. <BR> | |||
Again gently mix the contents of the well by moving the plate in gentle circles.<BR> | |||
<BR> | |||
At the end of these transfers, measure the optical density in each well using the plate reader. Then put your 96-well plate in a plastic tupperware container at 37C to grow. Take measurements on the optical density daily to confirm growth inhibition or stimulation. | |||
==Antibiotic Production== | |||
Many microbes secrete antimicrobial compounds to help them compete with other microorganisms for habitat. Some of the bacteria that are common antibiotic producers are the Actinomycetes (including ''Streptomycetes'' species), many of the ''Bacillus'' species, and the fruiting myxobacteria, to name just a few among many, many antibiotic producing bacteria. You can also test for the opposite: the sensitivity of your soil organisms (or known stock bacteria) to manufactured or secreted antibiotics.<BR> <BR> | |||
(This testing will take 3 weeks.) <BR> | |||
''' Week 1:''' <BR> | |||
Identify how many potential antibiotic producers you might have. Definitely test any isolates that are likely to be ''Actinomycetes'', ''Myxobacteria'', or ''Bacillus''. It's possible that you might discover the next great antimicrobial drug and get very rich by selling the patent for your discovery to a drug company. Remember that the discovery of penicillin was completely accidental. <BR><BR> | |||
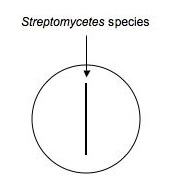
Using aseptic technique, transfer an isolated colony (possible ''Streptomyces,'' etc.) that's likely to be an antibiotic '''producer''' to a microfuge tube containing 500μL of sterile water. Vortex to mix. Using a sterile swab, dip the swab in the diluted bacteria and make an inoculation (as shown below) down the middle of a plate of nutrient agar. Make a second plate exactly like the first for each isolate to be tested. Label them carefully and incubate the plates for ~1 week at RT.<BR><BR> | |||
[[Image:strep1a.jpg]]<BR> | |||
'''Week 2'''<BR> | |||
TO BE PROVIDED in LAB 7:<BR> | |||
3 fresh cultures of ''Eschericia coli'' (Gram negative), ''Staphylococcus aureus'' (Gram positive) and grown in nutrient broth.**Make sure you tell your instructors when you will need these cultures!**<BR> | |||
'''PROTOCOL'''<BR><BR> | |||
'''Use the plate(s) from week 1<UL><LI> | |||
Use a sterile swab and aseptically apply a line of inoculation of each of the provided broth cultures of : ''E. coli'' and ''S. aureus''. | |||
as shown below. Draw a line perpendicular to the antibiotic producer's (''Streptomyces'') inoculation. Be careful not to touch the antibiotic producer's growth. Using the same swabs, inoculate a new NA plate (one plate for all three cultures)by making a line across the plate for ''E. coli and S. aureus''. Incubate this plate along with your test plate. It will serve as a control to make sure that lack of growth is due to antibiotic sensitivity and not to no living cells in the inoculum.<LI> | |||
[[Image:strep2.jpg]] | |||
<LI> | |||
<LI> | |||
Incubate for another week.</LI></UL> | |||
'''Week 3 <BR>''' | |||
<UL><LI> | |||
Examine the plate and look for evidence of inhibition of growth of "test" organisms near the antibiotic producer's midline streak. <LI> | |||
Draw the results and evaluate whether or not there was evidence that an antibiotic was produced by the organism and, if so, which of the bacteria tested were sensitive to it and to what degree. If you found no inhibition of growth, does that mean that your potential antibiotic producer does not secrete an any antimicrobial compounds? Why or why not? </LI></UL> | |||
<div class=noprint> | |||
==Biofilm Assay== | ==Biofilm Assay== | ||
| Line 14: | Line 101: | ||
"Protocol" | "Protocol" | ||
* The day before, create an overnight culture of your isolates to be tested by inoculating a single colony into a tube of nutrient | * The day before, create an overnight culture of your isolates to be tested by inoculating a single colony into a tube of nutrient broth (or other appropriate medium). | ||
* In the morning, measure tho OD of the culture and dilute to 0.05. Use this dilution to inoculate at least 3 wells in your polystyrene dish (add ~5 ml of media to each well). Include a medium only control. | * In the morning, measure tho OD of the culture and dilute to 0.05. Use this dilution to inoculate at least 3 wells in your polystyrene dish (add ~5 ml of media to each well). Include a medium only control. | ||
* Incubate for 24-48 hours at | * Incubate for 24-48 hours at 37°C, monitoring turbidity and growth daily. | ||
* At the completion of incubation, aspirate the liquid from the wells using a pasteur pipette. | * At the completion of incubation, aspirate the liquid from the wells using a pasteur pipette. | ||
* Wash the wells once with deionized, distilled water. | * Wash the wells once with deionized, distilled water. | ||
| Line 27: | Line 114: | ||
<br> | <br> | ||
==Quorum Sensing== | ==Bacterial Quorum Sensing== | ||
'''Quorum sensing - chemical signaling within our community'''<br> | |||
Many bacteria are able to secrete signals into their environment to sense their density. Since bacteria are single-celled organisms, why would it be important for them to sense density? A very well studied example of a quorum sensing system was discovered in ''Vibrio fisheri'', a bacterium that produces light only at high densities. Because the light produced by a single bacterium is unlikely to be detectable, it makes sense to wait until a "quorum" is reached before turning on the expensive metabolic pathway that creates light. In this way, a gene regulatory network is actually controlled by cell density. To hear more about it from another source, visit this YouTube video: [http://www.youtube.com/watch?v=1NoxOs-hcRU Bonnie Bassler]. <br> | |||
Today we will be setting up a test to see if any of your isolates are secreting an Auto-Inducer (AI) into the surrounding media. Specifically, we will be using a strain of bacterium called ''Chromobacterium violaceum''. This organism is a Gram-negative coccobacillus and a facultative anaerobe that is normally found in the soil. It produces a very strong purple pigment (hence the name) in response to AI. The response, a pigment called violacein, may be useful for the treatment of colon and other cancers. ''C. violaceum'' grows readily on nutrient agar, producing distinctive smooth low convex colonies with a dark violet metallic sheen. Its full genome was published in 2003. <BR> We will also use a violacein-negative, mini-Tn5 mutant of ''C. violaceum'' (CV026). This mutant can produce pigment in response to the AI from other bacteria, but can no longer secrete its own AI - this will be our biosensor. <br> | |||
1. You should have prepared for this assay by making a recent subculture streak plate of each of your isolates on Nutrient agar. These fresh sub-cultures should be no more than 1 week old when starting this test (unless an isolate that you want to test is a very slow grower). Throughout this procedure be sure to use good aseptic transfer technique. The heat stress of a hot loop can cause the transposon in the mutant strain to be lost from the cells so we will use either a plastic disposable sterile loop or a sterile toothpick instead of your wire loop for the inoculation of the mutant ''Chromobacterium''. <BR><BR> | |||
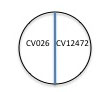
2. Use a marker to draw a line on the bottom of the plate dividing the plate into two halves. <BR> | |||
[[Image:Quorum1.jpg]]<BR> | |||
3. Label one side of the center line on a plate of nutrient agar with the name of the positive control bacterium, ''Chromobacterium violaceum ATCC 12472'' (the parent strain to CV026) and label CV026 mutant on the other side of the line. <br> | |||
[[Image:Quorum2a.jpg]]<BR> | |||
4. On all the other plates (one for each isolate to be tested), label CV026 mutant on one side and your isolate code name on the other side of the line. <BR> | |||
[[Image:Quorum3a.jpg]]<BR> | |||
<BR> | |||
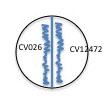
5. Begin with the young culture of CV026 mutant. Use the same sterile plastic loop to inoculate the CVO26 mutant on the appropriate side of ALL of your plates. Streak the inoculum near but not touching the center line (see illustration below). <br> | |||
[[Image:Quorum4a.jpg]]<BR> | |||
6. Use your heat sterilized and cooled wire loop to inoculate the appropriate side of the control plate that you labeled in step 3 using a fresh culture of ''C. violaceum'' 12472.<br> | |||
[[Image:Quorum6a.jpg]] [[Image:Quorum7a.jpg]]<BR> | |||
7. Use your wire loop, making sure it cools after flaming, to streak each of your isolates on the appropriate side of a pre-labeled plate, near but not touching the center line.<BR> | |||
[[Image:Quorum5a.jpg]]<BR> | |||
8. Incubate your cultures agar side up at RT for a week. If the isolate produces AI and it is sensed by the CV026 mutant, a purple pigment will be produced by the mutant. What does this tell you about your isolate? What are the kind of autoinducers that these bacteria might produce and sense?<br> | |||
Latest revision as of 08:39, 18 February 2013
Antagonistic and Mutualistic Interactions
*NOTE: You must remember to set up fresh nutrient broth cultures for your isolates 1-3 days before lab to do this test!
The microbial community living in your environments is a complex one with many different microorganisms. As is true of any environment, these microbes interact with each other - both functionally and physically. Do selected bacteria from your community help each other or harm each other while trying to find a niche in your community? Today, you will try to answer that question by testing your cultured isolates for examples of mutualism or antagonism (co-operation or competition)by culturing them in controlled communities. Some of these bacteria may prevent the growth of others through the production of chemical inhibitors; others might promote the growth of their neighbors by producing metabolites that are needed. We are going to look for both positive and negative interactions.
PREPARING THE ISOLATES:
You will inoculate 50 µl of log phase (young culture) isolate grown in fresh nutrient broth into the assigned well(s). Once again try to control for similar numbers of organisms in your inoculum by starting with normalized optical density measurements (start with a concentration of 0.5 OD600)
Interaction Assay Set Up
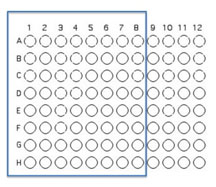
You will use 64 of the wells on a 96 well plate for this assay. Each pair will use 8 unique isolates to test for interactions. Use Excel to keep track of which organisms that will be inoculated into each well as described and illustrated below.
FOLLOW THE TEMPLATE CAREFULLY!!!!!! It is easy to get this inoculation messed up, but don't!

Transfer 50 μL of each of 8 unique isolates to be tested into the illustrated row of wells (A1 is Isolate 1, A2 is Isolate #2 etc through A8)
Beginning with Isolate #2, inoculate a second 50 μl of each of your isolates into the column wells B1, C1, etc. (indicated by the green color).
Add 100 μL of nutrient broth to each of the wells containing your isolates (row wells A1-A8 and column wells B1-H1)
Gently move the 96 well plate in a circular motion to mix.

Transfer 10 μl of the contents of the 7 wells - A2 (containing isolate #2 etc) through A8 to the empty wells in each column as indicated by the yellow arrows. You will need to remove the first tip from a multichannel pipette. If you are using the multichannel pipette, be sure that you work slowly and check that each pipette tip is evenly filled. You may need to tighten the tips by hand, if so be sure to only touch the part of the tip that sits on the multichannel pipette, you wouldn't want to contaminate your wells with human organsisms!
Transfer 10 μl of the contents of wells A1 (containing isolate #1 etc) through H1 to each well in the row as indicated by the red arrows.
Again gently mix the contents of the well by moving the plate in gentle circles.
At the end of these transfers, measure the optical density in each well using the plate reader. Then put your 96-well plate in a plastic tupperware container at 37C to grow. Take measurements on the optical density daily to confirm growth inhibition or stimulation.
Antibiotic Production
Many microbes secrete antimicrobial compounds to help them compete with other microorganisms for habitat. Some of the bacteria that are common antibiotic producers are the Actinomycetes (including Streptomycetes species), many of the Bacillus species, and the fruiting myxobacteria, to name just a few among many, many antibiotic producing bacteria. You can also test for the opposite: the sensitivity of your soil organisms (or known stock bacteria) to manufactured or secreted antibiotics.
(This testing will take 3 weeks.)
Week 1:
Identify how many potential antibiotic producers you might have. Definitely test any isolates that are likely to be Actinomycetes, Myxobacteria, or Bacillus. It's possible that you might discover the next great antimicrobial drug and get very rich by selling the patent for your discovery to a drug company. Remember that the discovery of penicillin was completely accidental.
Using aseptic technique, transfer an isolated colony (possible Streptomyces, etc.) that's likely to be an antibiotic producer to a microfuge tube containing 500μL of sterile water. Vortex to mix. Using a sterile swab, dip the swab in the diluted bacteria and make an inoculation (as shown below) down the middle of a plate of nutrient agar. Make a second plate exactly like the first for each isolate to be tested. Label them carefully and incubate the plates for ~1 week at RT.
Week 2
TO BE PROVIDED in LAB 7:
3 fresh cultures of Eschericia coli (Gram negative), Staphylococcus aureus (Gram positive) and grown in nutrient broth.**Make sure you tell your instructors when you will need these cultures!**
PROTOCOL
- Use a sterile swab and aseptically apply a line of inoculation of each of the provided broth cultures of : E. coli and S. aureus. as shown below. Draw a line perpendicular to the antibiotic producer's (Streptomyces) inoculation. Be careful not to touch the antibiotic producer's growth. Using the same swabs, inoculate a new NA plate (one plate for all three cultures)by making a line across the plate for E. coli and S. aureus. Incubate this plate along with your test plate. It will serve as a control to make sure that lack of growth is due to antibiotic sensitivity and not to no living cells in the inoculum.
-

- Incubate for another week.
Week 3
- Examine the plate and look for evidence of inhibition of growth of "test" organisms near the antibiotic producer's midline streak.
- Draw the results and evaluate whether or not there was evidence that an antibiotic was produced by the organism and, if so, which of the bacteria tested were sensitive to it and to what degree. If you found no inhibition of growth, does that mean that your potential antibiotic producer does not secrete an any antimicrobial compounds? Why or why not?