Koch Lab:Research: Difference between revisions
(New page: ==Coming soon==) |
No edit summary |
||
| Line 1: | Line 1: | ||
== | {{Koch_Lab}} | ||
<div style="padding: 10px; width: 720px; border: 5px solid #008;"> | |||
Our lab at UNM was started in August 2006, and full-time students will start in Summer of 2007. So as of March 2007, we are mostly cooking up ideas, ordering equipment and supplies, and starting to build some instrumentation. But we can generally describe our biophysics expertise that we intend to leverage and further develop as well as the general biological problems that we have learned a lot about and are planning to address with our collaborators. | |||
==Some of the biological problems we are actively interested in== | |||
Repair of [http://en.wikipedia.org/wiki/Dna_repair#Double-strand_breaks double-strand breaks] in DNA, [http://en.wikipedia.org/wiki/Chromatin chromatin] remodeling during [http://en.wikipedia.org/wiki/Transcription_%28genetics%29 gene transcription] (both of these are very important to genome stability and the development and progression of cancer), [http://en.wikipedia.org/wiki/RNA_polymerase_II RNA Polymerase II] elongation, site-specific protein-DNA interactions and binding kinetics, [http://en.wikipedia.org/wiki/Cytokinesis cytokinesis] and [http://en.wikipedia.org/wiki/Mitosis mitosis]. More details coming. | |||
==Some of our biophysics techniques== | |||
===Optical Tweezers=== | |||
[[Image:Unzipping DNA with Protein.png|right|thumb|Probing protein-DNA interactions by unzipping single DNA molecules with optical tweezers.]] | |||
Optical tweezers are formed by shining laser light into a microscope objective. The laser light can trap and move micron-sized particles (such as polystyrene microspheres) which are easily attached to biomolecules (such as DNA). A carefully constructed and calibrated optical tweezers system can position particles with sub-nanometer precision and apply forces over 100 piconewtons (which is enough to unravel a single double-stranded DNA molecule and break most non-covalent bonds). A very good description of optical tweezers can be found at the [http://www2.physics.umd.edu/~alaporta/ALPTech.html La Porta Lab website]. Optical tweezers now enable a wide array of fascinating experiments that provide critical information about biomolecular systems (See [http://www.stanford.edu/group/blocklab/ Steve Block Lab] for some famous examples with Kinesin and RNA Polymerase). To the right is a diagram of using optical tweezers to unzip single DNA molecules when there are DNA-binding proteins present in the solution. Our lab has expertise in this technique (see PMID 12124289) which we will use to answer key biological questions such as in chromatin remodeling during DNA damage repair. | |||
<br style="clear:both;"/> | |||
===Magnetic Tweezers=== | |||
[[Image:Mag Tweezers EWD.png|right|thumb|Schematic of magnetic tweezers apparatus under construction at CINT with P. Goodwin, J. Werner, D. Keller, G. Thayer, J. Martin, G. Bachand.]] | |||
In contrast to optical tweezers, the magnetic "tweezers" that we use are not really tweezers per se. They are really apparatuses that can apply calibrated, uni-directional, dynamic forces to many single-molecule tethers simultaneously. For example, the picture to the right shows an apparatus designed to apply vertical forces to many single-molecule tethers at once. Tether length information is provided by evanescent wave scattering (as the [http://www.physics.ucla.edu/zocchi_lab/ Zocchi lab] does), but there is no control over microsphere position...only applied force. The idea is to be able to perform experiments more rapidly, but with less exquisite control. Furthermore, magnetic experiments are much more amenable to varying the temperature and for samples containing a lot of debris that would interfere with optical traps. An example of an experiment we would like to perform is to set up single-molecule tethers in an unzipping configuration and then detect whether or not a DNA-binding protein is present based on the unzipping force. Because of the increased efficiency and robustness compared with optical tweezers, it would be much easier to probe how protein binding changes when the osmotic pressure, temperature, and other factors are varied. | |||
<br style="clear:both;"/> | |||
===MEMS and Nanofabricated Devices=== | |||
[[Image:PN_186_sensor_45deg_08a_big.PNG|right|thumb|SEM image of a MEMS force sensor described in our [http://link.aip.org/link/?APL/89/173901 APL paper]]] | |||
Our lab also has interest in developing micro- and nano-fabricated devices that can enable biophysical experiments that are impossible or extremely difficult with other methods. In principle, microfabricated experiment platforms can be more versatile, much more robust, and more widely accessible than optical tweezers setups. However, there is a very long way to go towards this goal. In collaboration with Maarten de Boer, Alex Corwin, Gayle Thayer, and George Bachand at Sandia, we have recently taken initial steps towards using MEMS devices for real biophysics experiments (see picture to the right). The device we used was fabricated with Sandia's SUMMiT process (via the [http://mems.sandia.gov/samples/index.html SAMPLES] program, and so is easily incorporated into other MEMS designs including actuation (comb drive or inch worm) and detection. Also, our lab is now located at the Center for High Technology Materials ([http://www.chtm.unm.edu CHTM]) which houses one of the [http://www.nnin.org/nnin_newmexico.html NNIN nanofabrication facilities]. We are working on ideas for leveraging the unique capabilities at the CHTM (for example, nanochannels fabricated via optical interference lithography over square-centimeter areas) for biological experiments, partucularly to complement or combine with the biological studies being performed with optical and magnetic tweezers. | |||
<br style="clear:both;"/> | |||
===Tethered Particle Motion=== | |||
A prerequisite for most our optical and magnetic tweezers experiments is to tether microspheres to a coverglass via a single-molecule (usually DNA). When viewed under a microscope, tethered microspheres (approximately 1 micron) undergo constrained Brownian motion. An expert at tethering can tell by eye whether the "amount" of Brownian motion is correct for the length and nature of the single-molecule tethers. It turns out that even without applying and measuring forces with optical or magnetic tweezers, useful biological experiments can be carried out by video microscopic analysis of the tethered particle motion (TPM). A famous early example of this was published by the [http://www.bact.wisc.edu/landick/ Landick Lab] in 1991: PMID 1861724. The hardware requirements for this method are only a microscope, video camera, and computer, making the method very appropriate to carry out in any biology lab with standard microscopy facilities. However, there is still a barrier presented by data acquisition and data analysis software. Our lab has expertise using LabVIEW for this application and we intend to both carry out our own TPM studies (starting with chromatin remodeling during double-strand break repair) as well as share our LabVIEW software and methods with other labs who can utilize the technique. | |||
===Simulation, Analysis, Modeling, Automation=== | |||
Our lab has expertise (mostly in LabVIEW) with software applications for a variety of purposes, as is common with experimental biophysics labs. Software for data acquisition and data analysis are critical for all of the above techniques. Furthermore, for many of the biological systems we are interested in, it comes in very handy to create simple models for the kinetic processes and then to perform [http://en.wikipedia.org/wiki/Monte_Carlo_method Monte Carlo simulations] to help guide our thinking in both designing and interpreting experiments. Currently, most of our software is only available in house, but with some work, we could make the software useful to other labs around the world (particularly those using LabVIEW). The more effort we put into making user-friendly software and also explaining all of the code, the more likely it is to be useful. Any inquiries from potential end users (see our [[Koch_Lab:Contact|contact page]]) would likely be very useful for spurring us on in these endeavors. Some possibly useful software: | |||
* Optical tweezers feedback control LabVIEW application. | |||
* Dynamic Force Spectroscopy (DFS) analysis software (would require data file interpreter). Also DFS simulation software. | |||
* Top-level application for simulation of enzymatic processes on chromatin (such as Pol II transcription) coupled with simulted Chromatin Immunoprecipitation ([http://en.wikipedia.org/wiki/Chromatin_immunoprecipitation ChIP]) analysis. Useful for evaluating alternative interpretations of sometimes-compliated ChIP experiments. | |||
* Tethered Particle Motion (TPM) software. | |||
==Some of the other labs that we talk with and may collaborate with== | |||
[http://hsc.unm.edu/som/micro/Maryann.html Mary Ann Osley Lab], UNM, HSC Department of Molecular Genetics and Microbiology<br> | |||
[http://dir.niehs.nih.gov/dirlmc/transcript.htm Karen Adelman Lab], NIH, NIEHS<br> | |||
[http://mcdb.omrf.org/gorbskylab/ Gary Gorbsky Lab], Oklahoma Medical Research Foundation<br> | |||
[http://cint.lanl.gov/user_call/scientist_summaries.shtml George Bachand Lab], Sandia National Labs and CINT<br> | |||
Alex Corwin, Maarten de Boer, and Gayle Thayer Labs, Sandia National Labs<br> | |||
[http://cint.lanl.gov/user_call/scientist_summaries.shtml Peter Goodwin & Jim Werner Labs], Los Alamos National Lab and CINT<br> | |||
Kim Rasmussen Lab, Los Alamos National Lab | |||
</div> | |||
Revision as of 10:19, 27 March 2007
| Home | Research | Lab Members | Publications | Protocols | Contact | Funding |
| Principles | Data | Notebooks | Links | Meetings | Presentations | Inventory |
Our lab at UNM was started in August 2006, and full-time students will start in Summer of 2007. So as of March 2007, we are mostly cooking up ideas, ordering equipment and supplies, and starting to build some instrumentation. But we can generally describe our biophysics expertise that we intend to leverage and further develop as well as the general biological problems that we have learned a lot about and are planning to address with our collaborators.
Some of the biological problems we are actively interested in
Repair of double-strand breaks in DNA, chromatin remodeling during gene transcription (both of these are very important to genome stability and the development and progression of cancer), RNA Polymerase II elongation, site-specific protein-DNA interactions and binding kinetics, cytokinesis and mitosis. More details coming.
Some of our biophysics techniques
Optical Tweezers

Optical tweezers are formed by shining laser light into a microscope objective. The laser light can trap and move micron-sized particles (such as polystyrene microspheres) which are easily attached to biomolecules (such as DNA). A carefully constructed and calibrated optical tweezers system can position particles with sub-nanometer precision and apply forces over 100 piconewtons (which is enough to unravel a single double-stranded DNA molecule and break most non-covalent bonds). A very good description of optical tweezers can be found at the La Porta Lab website. Optical tweezers now enable a wide array of fascinating experiments that provide critical information about biomolecular systems (See Steve Block Lab for some famous examples with Kinesin and RNA Polymerase). To the right is a diagram of using optical tweezers to unzip single DNA molecules when there are DNA-binding proteins present in the solution. Our lab has expertise in this technique (see PMID 12124289) which we will use to answer key biological questions such as in chromatin remodeling during DNA damage repair.
Magnetic Tweezers
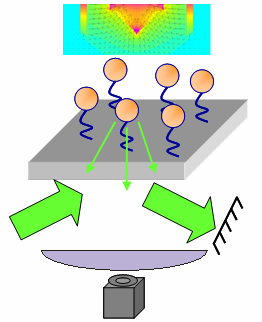
In contrast to optical tweezers, the magnetic "tweezers" that we use are not really tweezers per se. They are really apparatuses that can apply calibrated, uni-directional, dynamic forces to many single-molecule tethers simultaneously. For example, the picture to the right shows an apparatus designed to apply vertical forces to many single-molecule tethers at once. Tether length information is provided by evanescent wave scattering (as the Zocchi lab does), but there is no control over microsphere position...only applied force. The idea is to be able to perform experiments more rapidly, but with less exquisite control. Furthermore, magnetic experiments are much more amenable to varying the temperature and for samples containing a lot of debris that would interfere with optical traps. An example of an experiment we would like to perform is to set up single-molecule tethers in an unzipping configuration and then detect whether or not a DNA-binding protein is present based on the unzipping force. Because of the increased efficiency and robustness compared with optical tweezers, it would be much easier to probe how protein binding changes when the osmotic pressure, temperature, and other factors are varied.
MEMS and Nanofabricated Devices

Our lab also has interest in developing micro- and nano-fabricated devices that can enable biophysical experiments that are impossible or extremely difficult with other methods. In principle, microfabricated experiment platforms can be more versatile, much more robust, and more widely accessible than optical tweezers setups. However, there is a very long way to go towards this goal. In collaboration with Maarten de Boer, Alex Corwin, Gayle Thayer, and George Bachand at Sandia, we have recently taken initial steps towards using MEMS devices for real biophysics experiments (see picture to the right). The device we used was fabricated with Sandia's SUMMiT process (via the SAMPLES program, and so is easily incorporated into other MEMS designs including actuation (comb drive or inch worm) and detection. Also, our lab is now located at the Center for High Technology Materials (CHTM) which houses one of the NNIN nanofabrication facilities. We are working on ideas for leveraging the unique capabilities at the CHTM (for example, nanochannels fabricated via optical interference lithography over square-centimeter areas) for biological experiments, partucularly to complement or combine with the biological studies being performed with optical and magnetic tweezers.
Tethered Particle Motion
A prerequisite for most our optical and magnetic tweezers experiments is to tether microspheres to a coverglass via a single-molecule (usually DNA). When viewed under a microscope, tethered microspheres (approximately 1 micron) undergo constrained Brownian motion. An expert at tethering can tell by eye whether the "amount" of Brownian motion is correct for the length and nature of the single-molecule tethers. It turns out that even without applying and measuring forces with optical or magnetic tweezers, useful biological experiments can be carried out by video microscopic analysis of the tethered particle motion (TPM). A famous early example of this was published by the Landick Lab in 1991: PMID 1861724. The hardware requirements for this method are only a microscope, video camera, and computer, making the method very appropriate to carry out in any biology lab with standard microscopy facilities. However, there is still a barrier presented by data acquisition and data analysis software. Our lab has expertise using LabVIEW for this application and we intend to both carry out our own TPM studies (starting with chromatin remodeling during double-strand break repair) as well as share our LabVIEW software and methods with other labs who can utilize the technique.
Simulation, Analysis, Modeling, Automation
Our lab has expertise (mostly in LabVIEW) with software applications for a variety of purposes, as is common with experimental biophysics labs. Software for data acquisition and data analysis are critical for all of the above techniques. Furthermore, for many of the biological systems we are interested in, it comes in very handy to create simple models for the kinetic processes and then to perform Monte Carlo simulations to help guide our thinking in both designing and interpreting experiments. Currently, most of our software is only available in house, but with some work, we could make the software useful to other labs around the world (particularly those using LabVIEW). The more effort we put into making user-friendly software and also explaining all of the code, the more likely it is to be useful. Any inquiries from potential end users (see our contact page) would likely be very useful for spurring us on in these endeavors. Some possibly useful software:
- Optical tweezers feedback control LabVIEW application.
- Dynamic Force Spectroscopy (DFS) analysis software (would require data file interpreter). Also DFS simulation software.
- Top-level application for simulation of enzymatic processes on chromatin (such as Pol II transcription) coupled with simulted Chromatin Immunoprecipitation (ChIP) analysis. Useful for evaluating alternative interpretations of sometimes-compliated ChIP experiments.
- Tethered Particle Motion (TPM) software.
Some of the other labs that we talk with and may collaborate with
Mary Ann Osley Lab, UNM, HSC Department of Molecular Genetics and Microbiology
Karen Adelman Lab, NIH, NIEHS
Gary Gorbsky Lab, Oklahoma Medical Research Foundation
George Bachand Lab, Sandia National Labs and CINT
Alex Corwin, Maarten de Boer, and Gayle Thayer Labs, Sandia National Labs
Peter Goodwin & Jim Werner Labs, Los Alamos National Lab and CINT
Kim Rasmussen Lab, Los Alamos National Lab
