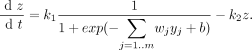Kara M Dismuke Week 10 Journal: Difference between revisions
From OpenWetWare
Jump to navigationJump to search
(→Equations: reformat bullets) |
(→Equations: equation 7 link) |
||
| Line 72: | Line 72: | ||
*{zc(t)}: reconstructed profile of z(t) in '''Z''' at all time points | *{zc(t)}: reconstructed profile of z(t) in '''Z''' at all time points | ||
EQUATION 7 <br> | EQUATION 7 <br> | ||
[[Image: Mml-math-7.gif]] | |||
*Linear form of the model | *Linear form of the model | ||
*parameters di (i=0,1,2): computed by minimizing error in function 6 | *parameters di (i=0,1,2): computed by minimizing error in function 6 | ||
Revision as of 20:22, 22 March 2015
Outline
Introduction
Regulation of gene expression
- important process in cell
- takes static information (in DNA) and transmits it into protein molecules (that serve various functions)
- requires recognition of specific promoter sequences
- the effects of transcription change with as the cell changes/develops
Microarrays
- document changes in gene expression over time
- analysis these changes can enable one to see a relationship between genes and their regulators
- use microarray data to track the interaction between genes and their regulators
Saccharomyces cerevisiae
- gene-expression data gathered from genome-wide microarrays
- data analyzed using clustering methods
- data modeled using singular value decomposition
- genes were grouped according to their transcriptional regulatory networks (i.e. relationship between the genes and their respective regulators/promoters)
Previous Studies
- use differential equations to try to develop a linear model that reflects the transcription pattern of each of the genes being studied
- Woolf and Wang: used "fuzzy logic" to try to do this
- Nachman: used kinetic model and Bayesian networks
- Bar-Joseph: used genomic information and analysis of gene expression data
- Wang and Makita: building of Bar-Joseph approach, the looked at the analysis of the promoter sequences and the sigma factor binding sequence motif
This Paper
- alternative method b/c uses a nonlinear differential equation model
- Procedure
- choose set of all potential regulators (chose pool of 184)
- choose set of target genes of S. cerevisiae (chose 40)
- picks genes from possible regulators and applies model to then compare results to information known about the target gene
- repeated to exhaust all possibilites
- determine which regulators correctly model gene expression model
- compare results and make conclusions using results from other studies & also a comparison of the linear model
- result: this method can correctly identify a target gene's specific regulator and can say whether or not that regulator is an activator or repressor
Results
Dynamic model of transcription control
Model's Assumptions
- recursive action of regulators on target gene (over time)
- regulatory effect on gene can be expressed with a combination of its regulators
Equations
- b: parameter that represents the initial delay or unspecific bias from regulatory effects associated with gene expression
- g: regulatory effect for particular gene
- wj: regulatory weights
- yj: expression level of regulators
- j=1,2,...m
- m: the number of regulators controlling the gene
- ρ: regulatory effects of other genes
- x: effect of degradation
- degradation: x = k*z where k is a constant in this kinetic equation
- ρ and x make up the rate of expression of a target gene (dz/dt)
- z: target expression level
- complete model for control of target gene expression z
- k1: maximal rate of expression
- k2: rate of degradation of target gene product
- simplification of Equation 3
- y: approximated with polynomial of degree n
- Once we have the expression profiles Z {z(t)} of the target and Y {y(t)} of the regulator genes, we search for gene profiles that minimize the mean square error function.
- t: 1,2,...Q
- Q: data points computed using Equation 4
- {zc(t)}: reconstructed profile of z(t) in Z at all time points
- Linear form of the model
- parameters di (i=0,1,2): computed by minimizing error in function 6
Definitions
- transcription
- Transcription is the first step of gene expression, in which a particular segment of DNA is copied into RNA by the enzyme RNA polymerase. Both RNA and DNA are nucleic acids, which use base pairs of nucleotides as a complementary language that can be converted back and forth from DNA to RNA by the action of the correct enzymes. During transcription, a DNA sequence is read by an RNA polymerase, which produces a complementary, antiparallel RNA strand called a primary transcript. As opposed to DNA replication, transcription results in an RNA complement that includes the nucleotide uracil (U) in all instances where thymine (T) would have occurred in a DNA complement. Also unlike DNA replication where DNA is synthesized, transcription does not involve an RNA primer to initiate RNA synthesis.Although Transcription is nice.
- http://www.biology-online.org/dictionary/Transcription
- RNA polymerase
- An enzyme that is responsible for making rna from a dna template. In all cells RNAP is needed for constructing rna chains from a dna template, a process termed transcription. In scientific terms, RNAP is a nucleotidyl transferase that polymerizes ribonucleotides at the 3' end of an rna transcript. Rna polymerase enzymes are essential and are found in all organisms, cells, and many viruses.
- http://www.biology-online.org/dictionary/RNA_polymerase
- promoter
- A site in a DNA molecule at which RNA polymerase and transcription factors bind to initiate transcription of mRNA.
- http://www.biology-online.org/dictionary/Promoter
- activator
- A DNA-binding transcription metabolite that positively modulates an allosteric Enzyme or regulates one or more genes by increasing the rate of transcription.
- http://www.biology-online.org/dictionary/Activator
- repressor
- A regulatory protein that binds to an operator and blocks transcription of the genes of an opreon
- http://www.biology-online.org/dictionary/Repressor
- regulator
- In genetics, a regulator pertains to a gene that codes for substances capable of repressing expression of another gene.
- http://www.biology-online.org/dictionary/Regulator
- mRNA
- Abbreviated form for messenger ribonucleic acid, the type of RNA that codes for the chemical blueprint for a protein (during protein synthesis).
- http://www.biology-online.org/dictionary/Mrna
- gene expression
- The conversion of the information from the gene into mRNA via transcription and then to protein via translation resulting in the phenotypic manifestation of the gene.
- http://www.biology-online.org/dictionary/Gene_Expression
- punative
- Denoting a supposition or inference based on what was commonly believed, reputed, or deemed rather than on a direct evidence
- http://www.biology-online.org/dictionary/Putative
- combinatorial
- Any system using a random assortment of components at any positions in the linear arrangement of atoms, i.e., a combinatorial library of mutations could contain positions where all four bases have been randomly inserted.
- http://www.biology-online.org/dictionary/Combinatorial