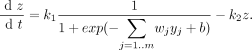Kara M Dismuke Week 10 Journal: Difference between revisions
From OpenWetWare
Jump to navigationJump to search
(→Conclusions: tables, figures) |
(update ppt) |
||
| (3 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
Journal Club 2 Group Presentation PPT: [[Media: Presentation_(1).pptx | JC2 PPT]] | |||
==Outline== | ==Outline== | ||
| Line 173: | Line 175: | ||
*B: activators have a similar curve when compared to that of the target gene and it's reconstructed profile | *B: activators have a similar curve when compared to that of the target gene and it's reconstructed profile | ||
**more activation, more gene expression (and less activation, less gene expression) | **more activation, more gene expression (and less activation, less gene expression) | ||
'''Figure 2''': Histogram of distribution of the order of correctly identified regulators in the sorted list of potential regulators (columns Min(m) and Min(m) from Table 1) | |||
*A: results from nonlinear model (equation 4) | *A: results from nonlinear model (equation 4) | ||
*B: results from linear model (equation 7) | *B: results from linear model (equation 7) | ||
*in comparing the two, one sees the nonlinear model better predicted the regulators of genes with a smaller pool (highest value for A was approximately between 19 and 21; highest value for B was approximately between 33 and 35) | *in comparing the two, one sees the nonlinear model better predicted the regulators of genes with a smaller pool (highest value for A was approximately between 19 and 21; highest value for B was approximately between 33 and 35) | ||
==Further Questions== | |||
What is the main result presented in this paper? | |||
*The main result presented in this paper was the discovery of a nonlinear method that can accurately pair regulators to their respective target genes, while also determining the function of the regulator (as a repressor or activator). | |||
What is the importance or significance of this work? | |||
*After reading this paper, it is clear there is no "end all be all" model that describes the relationships between regulators and their target genes. However, it became clear that their nonlinear model proved to be more effective in accurately identifying/describing these relationships than a linear model. In addition, the authors of the article noted the specificity of the case presented in the article and does not want their approach to eliminate all others; rather, they want them to work in conjunction with each other in order to develop a more full overall picture for the regulatory processes within yeast cells. | |||
The methods are described at points in the outline. | |||
*40 target genes | |||
*pool of 184 potential regulators | |||
*least square minimization used to help fit the data | |||
*for each target/regulator pair, the procedure was done 100 times with different initial conditions to obtain optimum fit | |||
==Definitions== | ==Definitions== | ||
Latest revision as of 23:59, 23 March 2015
Journal Club 2 Group Presentation PPT: JC2 PPT
Outline
Introduction
Regulation of gene expression
- important process in cell
- takes static information (in DNA) and transmits it into protein molecules (that serve various functions)
- requires recognition of specific promoter sequences
- the effects of transcription change with as the cell changes/develops
Microarrays
- document changes in gene expression over time
- analysis these changes can enable one to see a relationship between genes and their regulators
- use microarray data to track the interaction between genes and their regulators
Saccharomyces cerevisiae
- gene-expression data gathered from genome-wide microarrays
- data analyzed using clustering methods
- data modeled using singular value decomposition
- genes were grouped according to their transcriptional regulatory networks (i.e. relationship between the genes and their respective regulators/promoters)
Previous Studies
- use differential equations to try to develop a linear model that reflects the transcription pattern of each of the genes being studied
- Woolf and Wang: used "fuzzy logic" to try to do this
- Nachman: used kinetic model and Bayesian networks
- Bar-Joseph: used genomic information and analysis of gene expression data
- Wang and Makita: building of Bar-Joseph approach, the looked at the analysis of the promoter sequences and the sigma factor binding sequence motif
This Paper
- alternative method b/c uses a nonlinear differential equation model
- Procedure
- choose set of all potential regulators (chose pool of 184)
- choose set of target genes of S. cerevisiae (chose 40)
- picks genes from possible regulators and applies model to then compare results to information known about the target gene
- repeated to exhaust all possibilites
- determine which regulators correctly model gene expression model
- compare results and make conclusions using results from other studies & also a comparison of the linear model
- result: this method can correctly identify a target gene's specific regulator and can say whether or not that regulator is an activator or repressor
Results
Dynamic model of transcription control
Model's Assumptions
- recursive action of regulators on target gene (over time)
- regulatory effect on gene can be expressed with a combination of its regulators
Equations
- b: parameter that represents the initial delay or unspecific bias from regulatory effects associated with gene expression
- g: regulatory effect for particular gene
- wj: regulatory weights
- yj: expression level of regulators
- j=1,2,...m
- m: the number of regulators controlling the gene
- ρ: regulatory effects of other genes
- x: effect of degradation
- degradation: x = k*z where k is a constant in this kinetic equation
- ρ and x make up the rate of expression of a target gene (dz/dt)
- z: target expression level
- complete model for control of target gene expression z
- k1: maximal rate of expression
- k2: rate of degradation of target gene product
- simplification of Equation 3
- y: approximated with polynomial of degree n
- Once we have the expression profiles Z {z(t)} of the target and Y {y(t)} of the regulator genes, we search for gene profiles that minimize the mean square error function.
- t: 1,2,...Q
- Q: data points computed using Equation 4
- {zc(t)}: reconstructed profile of z(t) in Z at all time points
- Linear form of the model
- parameters di (i=0,1,2): computed by minimizing error in function 6
Computational Algorithm
- estimate expression profile of target gene in order to choose a set of potential regulators for a particular target gene
- search for potential regulators uses Equations 4 and 6
- approximate regulator gene profile by polynomial of degree n
- algorithm
- fit regulators using Equation 5
- choose target gene
- choose a regulatory gene from pool of possible regulators
- use least squares minimization on the target and regulator genes
- repeat for all possible regulators (step 3)
- choose regulators that best satisfy criterion
- repeat for all target genes (step two)
- procedure in algorithm was done 100 times for each pair of regulator and target gene
- optimization done using Levenberg-Marquardt procedure
- uses Runge-Kutta procedure (MATLAB's ode45 function)
Dataset selection
- evaluated model by using Spellman's dataset
- changes in gene expression: 18 time points over 2 cell cycle periods
- chip had 6178 open reading frames
- Spellman identified 800 genes associated with cell cycle, but in reality, there are a lot less regulators controlling the cell cycle
- this paper:
- 184 possible regulators (chosen based on YEASTRACT data and other papers' data
- chose 40 target genes (ones from Chen's paper)
Inference of Regulators
- data- in form of log base 2 of ratio between RNA amount divided by a standard
- prior to analysis, data was squared
- least squares minimization on each target gene for all possible regulators
- approximation of unknown real profile of a target gene (contains error)
- contains error, but this error can be estimated by this polynomial fit and/or a statistical model
- deviation from experimental data
- find "best" regulator for given target gene by finding regulator profile
- regulator profile- based on using model (equation 4) and minimizing E (equation 6)
Table 1: summary of 'correct identification' of regulators for all targets
- correct identification= gene identified as regulator for given target was also the regulator according to YEASTRACT
- note: YEASTRACT is a good resource, but still a work in progress
- correct identification- only 35% of the time, but false positive rate (FP) was very low
- FP rate: ratio between regulators identified as FPs and total number of potential regulators
- as criteria is softened, % found increased
- most increases can be attributed to a few targets (YOR323C, YJL155C, YDR285W, and YAL018C)
- specificity of the prediction: SP= (N-FP)/N
- N: number of potential regulators
- FP: number of false positives
- regulators either activate or repress the target gene
- classification was made based on the sign of the weight (w)
- algorithm correctly identified regulator as activator or repressor 75% of the time (based on YEASTRACT data)
Sources of Error
- YEASTRACT being incomplete
- experimental noise
- risk that the least squares minimization procedure does not yield optimal solution (although, there was attempt to avoid this by changing parameters and repeating the procedure 100 times for each target/regulator)
Comparison with linear model
- Linear model= Equation 7
- Figure 2: comparison between nonlinear model's and linear model's minimum for the number of regulators that needed to be tested before a yielding of the correct profile
- Table 1: nonlinear model fits better (by one order of magnitude) than linear model
- Regulators identified as "best fit" were compared against YEASTRACT and Chen's paper
- No matches with results in Chen's paper
- Nonlinear model prediction was the same as the linear model prediction in only 5 out of the 40 cases
Discussion
- create non-linear model that generates target gene expression profile from a specific regulator to help model the cell cycle
- minimize difference between measured target gene profile and profile computed from the regulator
- model can correctly identify regulators of target genes and determine their function as either an activator or repressor
- algorithm models all possible combinations of target/regulator and chooses from the pool of regulators to get the best predictions
- get complete information ability of each regulator to model each target gene profile so as to then determine the "best" regulator
- in comparison with the linear model, the nonlinear model gives much better results as it correctly identifies more regulators and gives a better fit of the computed target gene expression profile
- comparison between results from the Chen, nonlinear, and linear model, we get different results in terms of sets of genes
- focused on modeling simple case
- relies on outside knowledge (and if a regulator is not identified in outside sources, then it escapes being modeled)
- interactions between regulators and interactions between genes may skew results (not accounted for)
- models are designed for particular cases, and this model,was successful in capturing the behavior of transcriptional regulation with a fair amount of accuracy as well as identifying their function
- algorithm can be extended for use with a different organism's data set
- in future, model will be able to handle more complexity in the transcriptional regulatory interactions
Conclusions
- focus of this study: understand relationship between target genes and their regulators and understand the basic transcriptional regulation of the genes
- also, identify function of regulator as either an activator or repressor
- improvements to the algorithm can be made in the future to better account for the number of computations the algorithm requires
Figures and Tables
Table 1: "Summary of identification of regulators for 40 selected yeast cell cycle regulated genes"
- "best" column..."best" are regulators with smallest E
- as constraints on E are loosened, the number of regulators found/identified increases (in one case E1 is multiplied by 1.1 and in the other, E1 is multiplied by 1.2)
- min(m)...position of first correctly found regulator in list of regulators for given target
- two columns compare linear and non-linear model results
- E...E values
- two columns list lowest E values obtained from linear model and non-linear model
Figure 1
- 2 sections: A (graphs for regulators found that are repressors) and B (graphs for regulators found that are activators)
- x-axis: 18 time points used in model's simulation (to obtain data)
- y-axis: expression relative to time point zero
- names of target/regulator pair
- dotted line: reconstructed target (outputs from simulation)
- solid line: regulator that best fits the data for the specific target gene
- A: repressors have "opposite" curves to target gene and it's reconstructed profile
- more repression, less gene expression (and less repression, more gene expression)
- B: activators have a similar curve when compared to that of the target gene and it's reconstructed profile
- more activation, more gene expression (and less activation, less gene expression)
Figure 2: Histogram of distribution of the order of correctly identified regulators in the sorted list of potential regulators (columns Min(m) and Min(m) from Table 1)
- A: results from nonlinear model (equation 4)
- B: results from linear model (equation 7)
- in comparing the two, one sees the nonlinear model better predicted the regulators of genes with a smaller pool (highest value for A was approximately between 19 and 21; highest value for B was approximately between 33 and 35)
Further Questions
What is the main result presented in this paper?
- The main result presented in this paper was the discovery of a nonlinear method that can accurately pair regulators to their respective target genes, while also determining the function of the regulator (as a repressor or activator).
What is the importance or significance of this work?
- After reading this paper, it is clear there is no "end all be all" model that describes the relationships between regulators and their target genes. However, it became clear that their nonlinear model proved to be more effective in accurately identifying/describing these relationships than a linear model. In addition, the authors of the article noted the specificity of the case presented in the article and does not want their approach to eliminate all others; rather, they want them to work in conjunction with each other in order to develop a more full overall picture for the regulatory processes within yeast cells.
The methods are described at points in the outline.
- 40 target genes
- pool of 184 potential regulators
- least square minimization used to help fit the data
- for each target/regulator pair, the procedure was done 100 times with different initial conditions to obtain optimum fit
Definitions
- transcription
- Transcription is the first step of gene expression, in which a particular segment of DNA is copied into RNA by the enzyme RNA polymerase. Both RNA and DNA are nucleic acids, which use base pairs of nucleotides as a complementary language that can be converted back and forth from DNA to RNA by the action of the correct enzymes. During transcription, a DNA sequence is read by an RNA polymerase, which produces a complementary, antiparallel RNA strand called a primary transcript. As opposed to DNA replication, transcription results in an RNA complement that includes the nucleotide uracil (U) in all instances where thymine (T) would have occurred in a DNA complement. Also unlike DNA replication where DNA is synthesized, transcription does not involve an RNA primer to initiate RNA synthesis.Although Transcription is nice.
- http://www.biology-online.org/dictionary/Transcription
- RNA polymerase
- An enzyme that is responsible for making rna from a dna template. In all cells RNAP is needed for constructing rna chains from a dna template, a process termed transcription. In scientific terms, RNAP is a nucleotidyl transferase that polymerizes ribonucleotides at the 3' end of an rna transcript. Rna polymerase enzymes are essential and are found in all organisms, cells, and many viruses.
- http://www.biology-online.org/dictionary/RNA_polymerase
- promoter
- A site in a DNA molecule at which RNA polymerase and transcription factors bind to initiate transcription of mRNA.
- http://www.biology-online.org/dictionary/Promoter
- activator
- A DNA-binding transcription metabolite that positively modulates an allosteric Enzyme or regulates one or more genes by increasing the rate of transcription.
- http://www.biology-online.org/dictionary/Activator
- repressor
- A regulatory protein that binds to an operator and blocks transcription of the genes of an opreon
- http://www.biology-online.org/dictionary/Repressor
- regulator
- In genetics, a regulator pertains to a gene that codes for substances capable of repressing expression of another gene.
- http://www.biology-online.org/dictionary/Regulator
- mRNA
- Abbreviated form for messenger ribonucleic acid, the type of RNA that codes for the chemical blueprint for a protein (during protein synthesis).
- http://www.biology-online.org/dictionary/Mrna
- gene expression
- The conversion of the information from the gene into mRNA via transcription and then to protein via translation resulting in the phenotypic manifestation of the gene.
- http://www.biology-online.org/dictionary/Gene_Expression
- punative
- Denoting a supposition or inference based on what was commonly believed, reputed, or deemed rather than on a direct evidence
- http://www.biology-online.org/dictionary/Putative
- combinatorial
- Any system using a random assortment of components at any positions in the linear arrangement of atoms, i.e., a combinatorial library of mutations could contain positions where all four bases have been randomly inserted.
- http://www.biology-online.org/dictionary/Combinatorial