Julius B. Lucks/Antisense RNA Transcription Attenuation/Kinefold Results: Difference between revisions
No edit summary |
(→RNAI) |
||
| Line 44: | Line 44: | ||
T 62 80 4 | T 62 80 4 | ||
</pre> | </pre> | ||
[[Image:JBLucks_pT181_RNAI_BWF2B_pseudo.jpg]] | |||
[[Image:JBLucks_pT181_RNAI_BWF2B_no_pseudo.jpg]] | |||
Revision as of 20:14, 31 January 2008
Goal
Test wether or not the Kinefold RNA folding program produces the experimentally determined folds of RNAII/RNAIII. If this is true, then use it to identify a target region that we could mutate that would not drastically affect the secondary structure of RNAIII/II.
Kinefold is an RNA folding program that uses a kinetic, rather than a thermodynamic algorithm. It is thus able to identify kinetically trapped RNA folds, and able to incorporate pseudoknots. See http://kinefold.curie.fr for more information.
Initial Test
In (S Brantl, E G Wagner Mol Microbiol (2000) vol. 35 (6) pp. 1469-82), took sequence of RNAI (antisense RNA), and repC-RNA_132 (intermediate target RNA that looks like in the middle of the 2 hairpin, and the non-attenuateable fold. Folded in kinefold.
- both RNAI and RNAII affective at attenuation - RNAI shorter
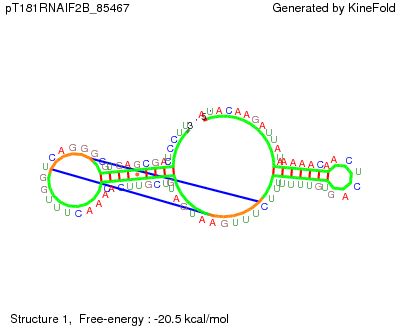
RNAI
- 85nt
- sequence: AUACAAGAUUAUAAAAACAACUCAGUGUUUUUUUCUUUGAAUGAUGUCGUUCACAAACUUUGGUCAGGGCGUGAGCGACUCCUU
Kinefold Request
Sequence name: pT181RNAIF2B Sequence : AUACAAGAUUAUAAAAACAACUCAGUGUUUUUUUCUUUGAAUGAUGUCGUUCACAAACUUUGGUCAGGGCGUGAGCGACUCCUU RNA sequence Type of Stochastic Simulation: Co-transcriptional Fold... A new base is added every 20.000000 milliseconds. No Simulated molecular time requested. Kinefold estimation : 2500 ms Pseudoknots: Allowed Entanglements: Non crossing Random seed: 97972 *********************************************************************** Actual sequence folded : AUACAAGAUUAUAAAAACAACUCAGUGUUUUUUUCUUUGAAUGAUGUCGUUCACAAACUUUGGUCAGGGCGUGAGCGACUCCUU Actual Traced and Forced Helices : T 46 79 9 T 13 32 7 T 38 53 6 T 35 69 6 T 62 80 4