In-fusion biobrick assembly: Difference between revisions
| Line 68: | Line 68: | ||
Ideally an assembly standard will use the same laboratory components (e.g. restriction enzymes and ligase) so that the same components can be used to assemble any two (or more) BioBricks together. This standard has one limitation in that custom primers need to be ordered for each individual assembly and hence requires different components. Nonetheless, this In-Fusion BioBrick Assembly method has been used to make several constructs and works well for myself and others in our lab. This method can be adapted to other plasmids in your favorite standard format and to assemble more than two BioBricks together. The method described here is a work-in-progress and the up-to-date method will be located here. Please document your success with using this method in the Notes section below! | Ideally an assembly standard will use the same laboratory components (e.g. restriction enzymes and ligase) so that the same components can be used to assemble any two (or more) BioBricks together. This standard has one limitation in that custom primers need to be ordered for each individual assembly and hence requires different components. Nonetheless, this In-Fusion BioBrick Assembly method has been used to make several constructs and works well for myself and others in our lab. This method can be adapted to other plasmids in your favorite standard format and to assemble more than two BioBricks together. The method described here is a work-in-progress and the up-to-date method will be located here. Please document your success with using this method in the Notes section below! | ||
==In-Fusion BioBrick Assembly Examples and Detailed Lab Protocols== | |||
[http://www.sys-bio.org/sbwWiki/_media/sysbio/labmembers/in-fusion_biobrick_assembly_022209.pdf In-Fusion BioBrick Assembly Lecture] | |||
[http://www.sys-bio.org/sbwWiki/_media/sysbio/labmembers/bioe498_lab_introduction_022409_3.pdf In-Fusion BioBrick Assembly Lab Introduction] | |||
[http://www.sys-bio.org/sbwWiki/_media/sysbio/labmembers/bioe498_lab1_022409_4.pdf In-Fusion BioBrick Assembly Lab 1] | |||
[http://www.sys-bio.org/sbwWiki/_media/sysbio/labmembers/bioe498_lab2_030309.pdf In-Fusion BioBrick Assembly Lab 2] | |||
[http://www.sys-bio.org/sbwWiki/_media/sysbio/labmembers/bioe498_lab3_030909_2.pdf In-Fusion BioBrick Assembly Lab 3] | |||
==Notes== | ==Notes== | ||
Revision as of 16:22, 21 May 2009
Overview
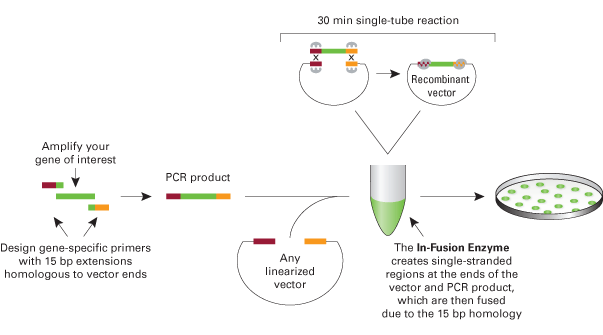
This is a method to assemble two BioBricks using the Clontech In-Fusion PCR Cloning Kit and maintains BioBrick standard formats. There are currently several BioBrick assembly standardsthat all involve assembly by restriction enzyme digestion and ligation. This method describes an alternative assembly method that allows for BioBricks to be assembled using the Clontech In-Fusion PCR Cloning Kit. One PCR-amplified BioBrick has homology on each end with the second PCR-amplified BioBrick (vector amplified with the BioBrick) to allow for the fragments to be fused together in the In-Fusion reaction (see figure below). This method can be adapted to fuse more than two BioBricks together and four fragments have been successfully fused. This method can also be used to re-engineer existing BioBricks by using mutagenic primers in the PCR reaction (see below for an example of simultaneous promoter and RBS re-engineering).
The advantages of In-Fusion BioBrick assembly over standard assembly are that it is faster, does not require restriction digestions or ligations or DNA extraction from a gel, and is more flexible in the sense that there is more control over the exact engineered sequence. The disadvantages are that the supplies are more expensive, custom primers are required, and occasionally there are mutations in assembled plasmids. However, sequencing a few miniprepped plasmids from positive colony PCR screened clones usually results in at least one plasmid without mutations.
Materials
- Thermocycler
- PCR tubes
- Primers
- Phusion PCR Mastermix(or another high fidelity polymerase mastermix)
- BioBrick template DNA (miniprepped plasmid diluted 1:1000 in water)
- Qiagen PCR Purification Kit
- Nanodrop (not required, but very useful)
- Clontech In-Fusion PCR Cloning Kit (dry down kit works well)
- TE Buffer (pH 8.0)
- Gel box, power supply, and gel supplies
- LB+Amp plates
- LB+Amp media
- Qiagen Miniprep Kit
Procedure
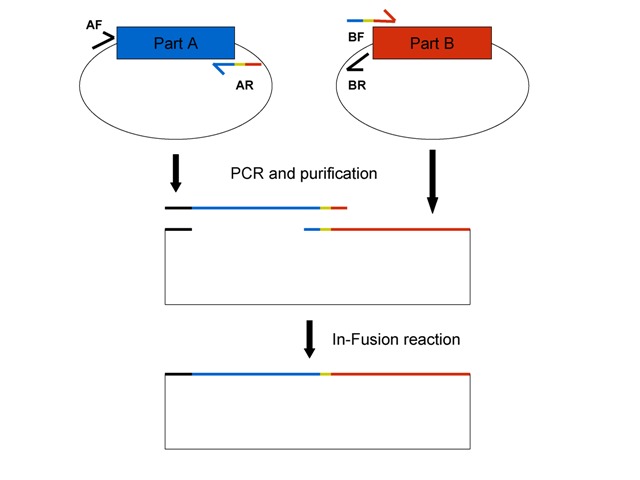
This figure shows the general In-Fusion BioBrick assembly scheme for BioBricks on pSB1A2 plasmids. See details below. Part A (blue) and Part B (red) are on pSB1A2 plasmids encoding ampicillin resistance. Primers described below are color-coded to show their homology. The thick black line indicates BioBrick prefix homology and the yellow sequence is the scar that is normally between parts after standard BioBrick assembly, if this is desired. If not, leave it off your primer (see below).
1. Read the In-Fusion manual and understand it completely before starting.
2. Order primers for parts to assemble. Primers can be optimized accordingly, but this general scheme should work for most BioBricks. The main rule is that the forward primer of one PCR-amplified fragment needs to have at least 15bp homology to the reverse primer of the second fragment, and vice versa, for the In-Fusion reaction to work. Also, it is better to put the vector (plasmid backbone) on the smaller BioBrick especially if the BioBrick is less than 100 bp since in my experience short fragments don't fuse together as well as larger fragments.
Primers for Part A (upstream part = insert) + Part B with the pSB1A2 vector (downstream part = vector):
- Part A Forward primer (AF): 5'-TTCTGGAATTCGCGGCCGCTTCTAG-3' (specific to the pSB1A2 prefix + 5 bases upstream of the prefix)
- Part A Reverse primer (AR): last 20 bases Part A + scar (if wanted) + first 20 bases Part B (reverse complement of this entire sequence)
- Part B + vector Forward primer (BF): reverse complement of Part A reverse primer
- Part B + vector reverse primer (BR): 5'-CTAGAAGCGGCCGCGAATTCCAGAA-3' (reverse complement of Part A forward primer)
Primers for Part A with the vector (upstream part = vector) + Part B (downstream part = insert):
- Part A + vector Forward primer: 5'-TACTAGTAGCGGCCGCTGCAGGCTTC-3' (specific to the pSB1A2 suffix + 5 bases downstream of the suffix)
- Part A + vector Reverse primer: last 20 bases Part A + scar (if wanted) + first 20 bases Part B (reverse complement)
- Part B Forward primer: reverse complement of Part A reverse primer
- Part B reverse primer: 5'-GAAGCCTGCAGCGGCCGCTACTAGTA-3' (reverse complement of Part A forward primer)
3. Perform a PCR reaction with a high fidelity polymerase such as Phusion using the recommended protocol. For template DNA, dilute miniprepped BioBrick plasmid DNA 1:1000 to minimize these “background” plasmids from getting transformed later. Digestion with DpnI after the PCR reaction may reduce background plasmids, but in my experience there is a high success rate (>50%) without the use of DpnI or phosphatase on the vector.
4. Run a gel with 5 ul of the purified PCR products to determine if the products are of the correct size and there are no secondary bands.
5. Purify PCR products with Qiagen PCR Purification Kit and elute with 30 ul water.
6. Measure the concentration of your PCR products with a Nanodrop or by comparing against a ladder on another gel.
7. Perform the In-Fusion reaction using the recommended protocol (this includes the transformation step with Fusion-Blue competent cells).
8. Perform colony PCR on at least 5 colonies (10-12 is ideal) using the VF2/VR primers or primers specific to your construct to find colonies that have the correct size insert.
9. Grow correct colony or colonies in LB+Amp overnight. Three is ideal since one should be correct without mutations which tend to occur most often at the junctions (regions of homology between fragments).
10. Perform miniprep(s) with the Qiagen Miniprep Kit.
11. Submit plasmid(s) for sequencing.
Discussion
Ideally an assembly standard will use the same laboratory components (e.g. restriction enzymes and ligase) so that the same components can be used to assemble any two (or more) BioBricks together. This standard has one limitation in that custom primers need to be ordered for each individual assembly and hence requires different components. Nonetheless, this In-Fusion BioBrick Assembly method has been used to make several constructs and works well for myself and others in our lab. This method can be adapted to other plasmids in your favorite standard format and to assemble more than two BioBricks together. The method described here is a work-in-progress and the up-to-date method will be located here. Please document your success with using this method in the Notes section below!
In-Fusion BioBrick Assembly Examples and Detailed Lab Protocols
In-Fusion BioBrick Assembly Lecture In-Fusion BioBrick Assembly Lab Introduction In-Fusion BioBrick Assembly Lab 1 In-Fusion BioBrick Assembly Lab 2 In-Fusion BioBrick Assembly Lab 3
Notes
Please feel free to post comments, questions, or improvements to this protocol. Happy to have your input!
- List troubleshooting tips here.
- You can also link to FAQs/tips provided by other sources such as the manufacturer or other websites.
- Anecdotal observations that might be of use to others can also be posted here.
Please sign your name to your note by adding '''*~~~~''': to the beginning of your tip.
References
Contact
sleight at u.washington.edu