IGEM:Paris Bettencourt 2012/Notebooks/MAGE group: Difference between revisions
(→SID) |
(→SID) |
||
| Line 423: | Line 423: | ||
*Colicin E2 (DNase activity, Tol-dependent) | *Colicin E2 (DNase activity, Tol-dependent) | ||
*Colicin D (RNase activity, TonB-dependent) | *Colicin D (RNase activity, TonB-dependent) | ||
[[Image:sid.png]] | |||
Revision as of 03:23, 8 August 2012
| Notebooks | Design | Roadmap | Meetings and to-dos | Protocols | Bibliography | Previous Biosafety iGEM projects |
|---|
Week 1 & 2
looking for MAGE protocols from past iGEM team
Harvard 2011
MAGE
MAGE oligo strand choice
Choosing the strand
MAGE is much more efficient if the lagging strand is targeted with the MAGE oligo. It is thought that the MAGE oligo takes the place of the RNA primer that begins Okazaki fragments on the lagging strand during DNA replication. The diagram below shows which strand to choose when designing MAGE oligos. Use ecocyc.org[2] to determine where on the genome your site of interest lies, and which strand it's on (determined by forward or reverse orientation in the genome browser). Examples
- For HisB deletion, we used the same strand, and for the rpoZ deletion, we used the reverse complement.
Protocol
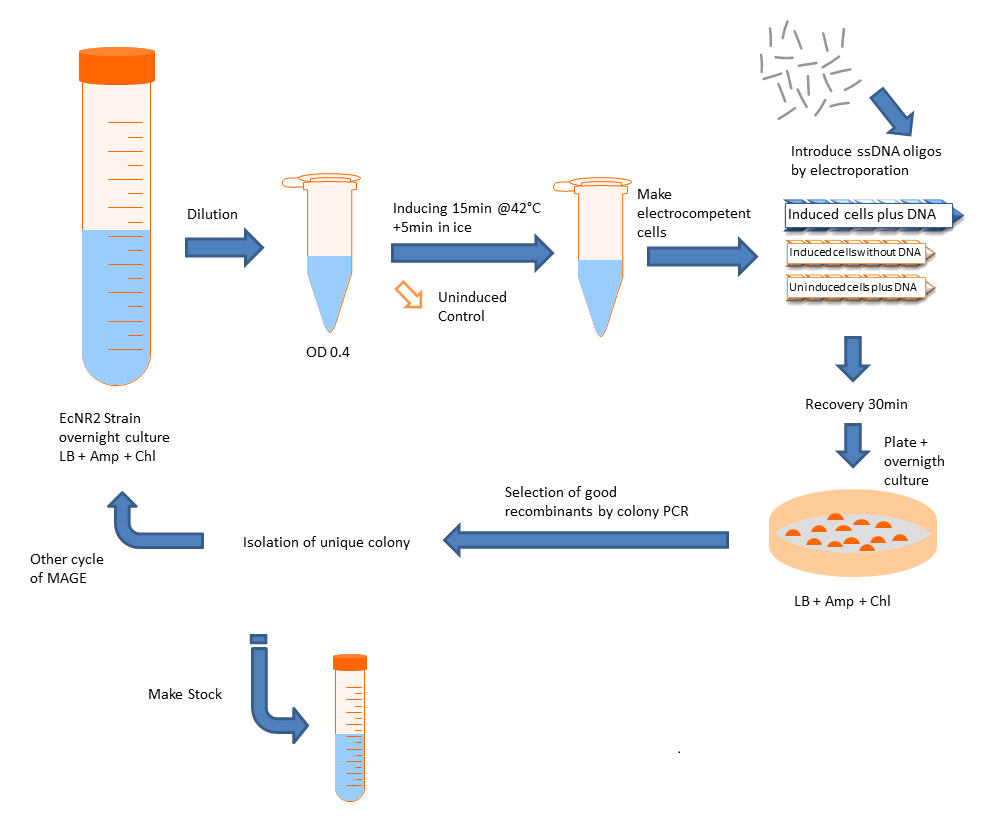
- Grow up cells to mid-log at 30C.
- Induce lambda red machinery by incubating cells at 42C for 15 minutes (for other strains, induction procedure may be different).
- Centrifuge 1-1.5mL of culture at 4C for 1 minute at top speed
- Remove all the media from the pellet and resuspend in 1mL of cold ddH2O before spinning again at 4C for 1 min at top speed.
- Repeat for a second water wash.
- Remove water carefully, taking care not to disturb the cell pellet. Add the MAGE oligo (amount will vary: add enough to have a final concentration of 2.5μM in a total volume of 50μL) and cold ddH2O to bring the volume up to 50μL. Mix and transfer to a cold, 1mm gap cuvette for electroporation.
-Note: MAGE oligo should preferentially be concentrated enough that only a few microliters need to be added. Adding larger amounts, especially if the salt concentration is high, may interfere with electroporation.
- Thoroughly dry the sides of the cuvette and electroporate at 1.80 kV for ideally 5.7 ms (Ec1 setting on many electroporators).
- Immediately add 1mL of plain LB to the cuvette, pipette up and down to mix, and transfer to a culture tube containing an additional 2mL of plain LB.
- Recover at 30C at least 1 hour before plating, or if performing additional rounds of MAGE, let culture grow until it reaches mid-log and repeat procedure.
Yale 2011
- "Received 22 degenerate oligonucleotide sequences from Keck and performed MAGE. Cultures were grown up to midlog post electroporation and freeze-thawed approximately 14 times before plating. Suriving colonies were isolated, and we will soon put them through another cycle of MAGE. The process will be repeated. "
List of Auxotrophic Strains
- Alanine There are multiple pathways for alanine biosynthesis, and no known complete auxotrophs exist.
- Arginine Knockout in argA (Strains JW2786-1 and NK5992)
- Asparagine Requires knockouts in both asnA and asnB (strain ER)
- Aspartic acid Requires knockouts in both aspC and tyrB (strain DL39)
- Cysteine Knockout in cysE (Strains JW3582-2 and JM15)
- Glutamic acid Requires knockouts in both gltB and gdhA (strain PA340)
- Glutamine Knockout in glnA (Strains JW3841-1 and M5004)
- Glycine Knockout in glyA (Strains JW2535-1 and AT2457)
- Histidine Knockout in hisB (Strains JW2004-1 and SB3930)
- Isoleucine Knockout in ilvA (Strain JW3745-2 and AB1255)
- Leucine Knockout in leuB (Strain JW5807-2 and CV514)
- Lysine Knockout in lysA (Strain JW2806-1 and KL334)
- Methionine Knockout in metA (Strain JW3973-1 and DL41)
- Phenylalanine Knockout in pheA (Strain JW2580-1 and KA197)
- Proline Knockout in proA (Strain JW0233-2 and NK5525)
- Serine Knockout in serA (Strain JW2880-1 and JC158)
- Threonine Knockout in thrC (Strain JW0003-2 and Gif 41)
- Tryptophan Knockout in trpC (Strain JW1254-2 and CAG18455)
- Tyrosine Knockout in tyrA (Strain JW2581-1 and N3087)
- Valine/Isoleucine/Leucine Knockout in ilvD (Strain JW5605-1 and CAG18431)
oligos
optMAGE
http://arep.med.harvard.edu/optMAGE/
Papers
http://www.pnas.org/content/98/12/6742.full
http://www.pnas.org/content/100/26/15748.full
http://www.sciencedirect.com/science/article/pii/S0022283611000593
Control
lacZ Replicore 1 G 365'242 -> C mistmatch C~C and stop codon
*90 oligo generated by optMAGE
GGCCGATACTGTCGTCGTCCCCTCAAACTGGCAGATGCACGGTTANGATGCGCCCATCTACACCAACGTGACCTATCCCATTACGGTCAA
*70 oligo generated by optMAGE
GTCGTCGTCCCCTCAAACTGGCAGATGCACGGTTANGATGCGCCCATCTACACCAACGTGACCTATCCCA
test
- λred genome or plasmid
- ΔmutS or mutS
- 90 bases oligo or 70 bases oligo
- phosporilated or not
- 90
lacZp1 111 113 + 1 M 0 -11.87 90 1 0 0 10.79 ACGCCAGGGTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGCgAATCCGTAATCATGGTCATTTGTCCCCTCTTTCCCGGGCGCTAT lacZp2 111 113 + 2 M 0 -7.56 90 1 0 0 12.61 GATAGCGCCCGGGAAAGAGGGGACAAATGACCATGATTACGGATTcGCTGGCCGTCGTTTTACAACGTCGTGACTGGGAAAACCCTGGCG lacZm1 111 113 - 1 M 0 -10.91 90 1 0 0 11.19 ACGCCAGGGTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGCcAATCCGTAATCATGGTCATTTGTCCCCTCTTTCCCGGGCGCTAT lacZm2 111 113 - 2 M 0 -7.1 90 1 0 0 12.81 GATAGCGCCCGGGAAAGAGGGGACAAATGACCATGATTACGGATTgGCTGGCCGTCGTTTTACAACGTCGTGACTGGGAAAACCCTGGCG
- 70
lacZp1 111 113 + 1 M 0 -7.02 70 1 0 0 7.91 TTTCCCAGTCACGACGTTGTAAAACGACGGCCAGCgAATCCGTAATCATGGTCATTTGTCCCCTCTTTCC lacZp2 111 113 + 2 M 0 -4.37 70 1 0 0 8.60 GGGAAAGAGGGGACAAATGACCATGATTACGGATTcGCTGGCCGTCGTTTTACAACGTCGTGACTGGGAA lacZm1 111 113 - 1 M 0 -7.2 70 1 0 0 7.86 TTTCCCAGTCACGACGTTGTAAAACGACGGCCAGCcAATCCGTAATCATGGTCATTTGTCCCCTCTTTCC lacZm2 111 113 - 2 M 0 -4.37 70 1 0 0 8.60 GGGAAAGAGGGGACAAATGACCATGATTACGGATTgGCTGGCCGTCGTTTTACAACGTCGTGACTGGGAA
Week 3&4
Order oligo from IDT
- Stock @-20°C 2µg/µL
- LacZ TAA 100 nmole DNA oligo
GGC CGA TAC TGT CGT CGT CCC CTC AAA CTG GCA GAT GCA CGG TTA AGA TGC GCC CAT CTA CAC CAA CGT GAC CTA TCC CAT TAC GGT CAA
- LacZ* TAA 100 nmole DNA oligo
G*G*C* C*GA TAC TGT CGT CGT CCC CTC AAA CTG GCA GAT GCA CGG TTA AGA TGC GCC CAT CTA CAC CAA CGT GAC CTA TCC CAT TAC GGT CAA
- RS1 100 nmole DNA oligo
ATA CGC ATA TTG TTC GTT TGG ATG GAA TAC CAT ATG ACG CGG GCC CGC CCC TTC AAC GGT GGT CAC TTC CGC AGG GTC CTG CGC CAC GAG
- RS2 100 nmole DNA oligo
AGA ACC AGC AAC CAC TAC GCC CGC TGC GCC CAG TTT CTT CAG GCC CGC CTG ATC CAC TTT AGA CAC ATC AGC AAC GCT GAC GCG CAG ACG
- RS3 100 nmole DNA oligo
TGA TAA ATA CCG GGG CCG GAT GGT GGC GCA CCG TCA CAC GGG CCG TCC GGA AAT CCG TTA CCG TTA CGA CAG CGA CGG GCG GGT GAC AGA
- RS4 100 nmole DNA oligo
TGA TAA ATA CCG GGG CCG GAT GGT GGC GCA CCG TCA CAC GGG CCG TCC GGA AAT CCG TTA CCG TTA CGA CAG CGA CGG GCG GGT GAC AGA
MAGE strains received
- Prepare plates with Amp and chloramphenicol
- Plate and growth overnight
- Glycerol stock
- Culture overnight
- Make Electrocompetent cells
Protocol
MAGE on LacZ
- skeleton :
- Results :
- attempt 001
- Cell only : Dilution 10^-6 182B 0W 5,70
- LacZ 100ng : 10^-6 282B 0W 5,70
- LacZ* 100ng : 10^-5 478B 3W 5,70
- LacZ* 100ng : X X 1,1 too much DNA
- LacZ 50ng : 10^-5 SS8B 2W 5,70
- LacZ 50ng : 10^-5 412B 1W 5,70
- attempt 002
- Cell only : Dilution 10^-5 19B 0W 5,70
- Cell uninduced + lacZ 100ng : Dilution 10^-6 2B 0W 5,70
- Cell uninduced + lacZ 100ng : Dilution 10^-5 445B 0W 5,70
- LacZ 100ng : 10^-6 7B 0W 5,70
- LacZ 100ng : 10^-5 57B 0W 5,70
- LacZ* 100ng : 10^-6 0B 0W 5,70
- LacZ* 100ng : 10^-5 13B 0W 5,70
- LacZ 50ng : 10^-6 5B 0W 5,70
- LacZ 50ng : 10^-5 48B 0W 5,70
- LacZ* 50ng : 10^-6 0B 0W 5,70
- LacZ* 50ng : 10^-5 13B 0W 5,70
- LacZ 25ng : 10^-6 3B 0W 5,70
- LacZ 25ng : 10^-5 13B 0W 5,70
- LacZ* 25ng : 10^-6 25 0W 5,70
- LacZ* 25ng : 10^-5 174B 0W 5,70
- attempt 003
- In process...
- attempt 001
MAGE on RS
- skeleton :
KEIO PyrF
- Got the JW1273 strain for PyrF deletion from Necker
- Put it in incubator at 37°C
- glycerol stock
- Do not grow on minimal media
Week 5&6
KO sessions
Designing necessary primers
Plasmids
- First look at the sequence of the plasmid containing the resistance marker you wish to swap in for your target gene.
pKD3 (CamR)
- priming site 1: GTGTAGGCTGGAGCTGCTTC
- priming site 2: GGACCATGGCTAATTCCCAT
- priming site 2 reverse complement: ATGGGAATTAGCCATGGTCC
pKD4
- priming site 1: GTGTAGGCTGGAGCTGCTTC
- priming site 2: GGACCATGGCTAATTCCCAT
- priming site 2 reverse complement: ATGGGAATTAGCCATGGTCC
pKD13
- priming site 1: GTGTAGGCTGGAGCTGCTTC
- priming site 4: TTAATTCTCATGTTTGACAG
- priming site 4 reverse complement: CTGTCAAACATGAGAATTAA
Genes
- Next, find the sequence (and context sequence) of the gene you wish to remove
rhsA
- NC_000913 - rhsA gene +50bp upstream +50bp downstream
AGGGTTTCGCCTGTCAGCAGACAAATAACCCGATAAAACAAGGATGAGAAATGAGCGGAAAACCGGCAGCGCGTCAGGGCGACATGACGCAGTATGGCGGTAGC ATTGTTCAGGGTTCAGCCGGGGTGCGCATTGGTGCCCCCACCGGCGTGGCCTGTTCGGTGTGCCCCGGCGGAGTGACGTCCGGCCATCCGGTCAATCCCCTGC TCGGTGCAAAGGTCCTTCCCGGTGAAACCGACATCGCCCTGCCCGGCCCGCTGCCGTTCATCCTCTCCCGCACCTACAGCAGTTACCGGACAAAAACGCCCGC GCCGGTGGGGAGCCTCGGCCCCGGCTGGAAAATGCCTGCGGATATCCGCTTACAGCTGCGCGATAACACACTGATACTCAGTGATAACGGCGGCAGAAGCCTG TATTTTGAGCACCTGTTTCCCGGTGAGGACGGTTACAGCCGCAGCGAGTCACTGTGGCTGGTGCGCGGCGGCGTGGCGAAACTGGATGAAGGTCACCGGCTGG CCGCACTCTGGCAGGCGCTGCCGGAAGAACTCCGCTTAAGTCCGCATCGTTATCTGGCGACAAACAGTCCGCAGGGGCCGTGGTGGCTGCTCGGTTGGTGTGA GCGGGTGCCGGAAGCGGATGAGGTGCTGCCTGCGCCGCTGCCGCCGTACCGGGTACTGACCGGGCTGGTGGACCGCTTCGGGCGCACACAGACGTTCCACCGC GAAGCCGCCGGTGAATTCAGCGGCGAAATCACCGGCGTGACGGATGGTGCCTGGCGTCACTTCCGGCTGGTACTGACCACGCAGGCGCAGCGGGCAGAAGAAG CCCGGCAGCAGGCCATTTCCGGCGGGACGGAACCGTCCGCTTTTCCTGATACCCTGCCGGGTTACACCGAATATGGCCGGGACAACGGCATCCGTCTGTCTGC CGTGTGGCTGACGCACGACCCGGAATACCCGGAGAATTTACCTGCCGCGCCGCTGGTGCGCTATGGCTGGACGCCACGCGGCGAACTGGCGGTGGTGTATGAC CGTAGTGGCAAACAGGTGCGCAGCTTTACTTACGATGATAAATACCGGGGCCGGATGGTGGCGCACCGTCACACGGGCCGGCCGGAAATCCGTTACCGTTACG ACAGCGACGGGCGGGTGACAGAACAGCTAAACCCGGCAGGCTTAAGCTACACGTATCAGTATGAGAAAGACCGCATCACCATCACCGACAGCCTGGACCGCCG TGAAGTGCTGCACACGCAGGGCGAAGCCGGGCTGAAGCGGGTGGTGAAAAAGGAACACGCGGACGGCAGCGTCACGCAGAGTCAGTTTGACGCCGTGGGCAGG CTCAGGGCACAGACGGATGCCGCAGGCAGGACAACAGAGTACAGCCCGGATGTGGTGACGGGCCTCATCACGCGCATAACCACGCCGGATGGCAGGGCATCGG CGTTTTACTATAACCACCACAACCAGTTAACGTCAGCCACCGGGCCTGACGGGCTGGAATTGCGCCGGGAATATGATGAATTGGGCCGTCTGATTCAGGAAAC TGCCCCTGACGGCGATATCACCCGCTACCGTTATGATAATCCACACAGTGACTTACCCTGCGCAACGGAAGATGCCACCGGCAGCCGGAAAACCATGACGTGG AGCCGTTACGGTCAGTTGCTGAGCTTCACCGACTGTTCCGGTTATGTAACCCGTTATGACCATGACCGCTTCGGGCAGATGACGGCGGTGCACCGCGAGGAAG GGCTGAGTCAGTACCGCGCATACGACAGCCGTGGACAGTTAATTGCCGTGAAAGACACGCAGGGCCATGAAACGCGGTATGAATACAACATCGCCGGTGACCT GACCGCCGTCATTGCCCCGGACGGCAGCAGAAACGGGACACAGTACGATGCGTGGGGAAAGGCCGTCCGTACCACGCAGGGCGGGCTAACGCGCAGTATGGAA TACGATGCTGCCGGACGGGTCATCCGCCTGACCAGTGAAAACGGCAGCCACACCACCTTCCGTTACGATGTACTTGACCGGCTGATACAGGAAACCGGCTTTG ACGGCCGCACACAGCGTTATCACCACGACCTGACCGGCAAACTTATCCGCAGCGAGGATGAGGGTCTGGTCACCCACTGGCACTATGACGAAGCAGACCGCCT CACGCACCGCACCGTGAAGGGTGAAACCGCAGAGCGGTGGCAGTATGACGAACGTGGCTGGCTGACAGACATCAGCCATATCAGCGAAGGGCACCGGGTGGCG GTGCATTACAGGTATGATGAGAAAGGCCGGCTGACCGGTGAGCGTCAGACGGTGCATCACCCGCAGACGGAAGCACTGCTCTGGCAGCATGAGACCAGACATG CGTACAACGCGCAGGGGCTGGCGAACCGCTGTATACCGGACAGCCTGCCCGCCGTGGAATGGCTGACCTACGGCAGCGGTTACCTGGCAGGCATGAAACTCGG CGACACACCGCTGGTGGAGTACACCCGCGACCGCCTGCACCGGGAAACGCTGCGCAGCTTCGGCCGTTATGAACTCACCACCGCTTATACCCCTGCCGGGCAG TTACAGAGCCAGCACCTGAACAGCCTGCTGTCTGACCGCGATTACACCTGGAACGACAACGGCGAACTCATCCGCATCAGCAGCCCGCGCCAGACCCGGAGTT ACAGCTACAGCACCACCGGCAGGCTGACCGGCGTTCACACCACCGCAGCGAATCTGGATATCCGCATCCCGTATGCCACAGACCCGGCAGGTAACCGCCTGCC CGACCCGGAGCTGCACCCGGACAGCACCCTCAGCATGTGGCCGGATAACCGTATCGCCCGTGACGCGCACTATCTTTACCGGTATGACCGTCACGGCAGGCTG ACAGAGAAAACCGACCTCATCCCGGAAGGGGTTATCCGCACGGATGATGAGCGGACTCACCGGTACCATTACGACAGTCAGCACCGGCTGGTGCACTACACGC GGACACAATATGAAGAGCCGCTGGTCGAAAGTCGCTATCTTTACGACCCGCTGGGCCGCAGGGTGGCAAAACGGGTGTGGCGGCGTGAACGGGACCTGACGGG CTGGATGTCGCTGTCACGGAAACCGCAAGTGACCTGGTACGGCTGGGACGGCGACCGGCTGACCACAATACAGAACGACAGAACCCGCATCCAGACGATTTAT CAGCCGGGGAGCTTCACGCCACTCATCAGAGTTGAAACCGCCACCGGTGAGCTGGCGAAAACGCAGCGCCGCAGCCTGGCGGATGCGCTTCAGCAGTCCGGCG GCGAAGACGGTGGCAGTGTGGTGTTCCCGCCGGTGCTGGTGCAGATGCTCGACCGGCTGGAAAGTGAAATCCTGGCTGACCGGGTGAGTGAGGAAAGCCGCCG CTGGCTGGCATCGTGCGGCCTGACCGTGGAGCAGATGCAAAACCAGATGGACCCGGTGTACACGCCGGCGCGAAAAATCCACCTGTACCACTGCGACCATCGC GGCCTGCCGCTGGCCCTTATCAGCAAGGAAGGGACAACAGAATGGTGCGCAGAATACGATGAATGGGGCAACCTGCTGAATGAAGAGAACCCGCATCAGCTGC AGCAGCTTATCCGCCTGCCGGGGCAGCAGTATGATGAGGAGTCCGGCCTGTATTACAACCGCCACCGCTATTATGACCCGCTGCAGGGGCGGTATATCACTCA GGATCCGATTGGGCTGAAGGGGGGATGGAATTTTTATCAGTATCCGTTGAATCCAGTTACGAATACAGATCCTCTGGGGTTAGAAGTTTTTCCTAGACCATTC CCCTTGCCAATTCCATGGCCCAAAAGCCCTGCACAGCAGCAAGCAGATGATAATGCTGCAAAAGCATTGACAAAATGGTGGAACGATACAGCATCACAAAGAA TATTTGACTCTCTAATATTGAATAATCCGGGACTAGCATTAGATATAACAATGATAGCTTCTCGTGGAAATGTTGCAGACACAGGGATAACTGATCGTGTCAA TGACATAATAAATGACAGATTCTGGAGTGATGGGAAAAAACCCGACAGATGTGACGTACTTCAGGAACTAATTGATTGTGGTGATATTAGTGCTAAAGATGCA AAAAGCACACAGAAAGCCTGGAATTGTCGTCACTCCAGACAGTCAAACGATAAAAAAAGATAGCCCTTGTGGAGGTTCCTGCAATGTCAAATACATACCAGAAAAGAAAGGCA
- Construct primers that have internal overlap with the resistance marker (pKD3/4) and external overlap with the target knockout gene.
- Forward primer: A
- AGGGTTTCGCCTGTCAGCAGACAAATAACCCGATAAAACAAGGATGAGAAGTGTAGGCTGGAGCTGCTTC
- Reverse primer: B
- TGCCTTTCTTTTCTGGTATGTATTTGACATTGCAGGAACCTCCACAAGGGATGGGAATTAGCCATGGTCC
- Forward primer: A
- Construct primers that only flank the target gene for PCR verification
- Forward primer: C
- CCAAAACAGCTTTCGCTACGTTGCT (25 bases)
- Reverse primer: D
- TGCCTTTCTTTTCTGGTATGTATTT (25 bases)
- Forward primer: C
rhsB
- NC_000913 - rhsB gene +50bp upstream +50bp downstream
CTTTTTCGTATGAAGATACTGTCATTAAAATAATAGAAAAGGATTTTACGATGAGCGGAAAACCGGCGGCGCGTCAGGGTGACATGACGCAGTATGGCGGTAGCATTGTTC AGGGTTCAGCCGGGGTACGTATTGGTGCCCCCACCGGCGTGGCCTGTTCGGTGTGCCCCGGCGGAGTGACGTCCGGCCATCCGGTCAATCCCCTGCTCGGTGCAAAGGTCCTT CCCGGTGAAACCGACATCGCCCTGCCCGGCCCGCTGCCGTTCATCCTCTCCCGCACCTACAGCAGTTACCGGACAAAAACGCCCGCGCCGGTGGGGAGCCTCGGCCCCGGCTG GAAAATGCCTGCGGATATCCGCTTACAGCTGCGCGATAACACACTGATACTCAGTGATAACGGCGGCAGAAGCCTGTATTTTGAGCACCTGTTTCCCGGTGAGGACGGTTACA GCCGCAGCGAGTCACTGTGGCTGGTGCGCGGCGGCGTGGCGAAACTGGATGAAGGTCACCGGCTGGCCGCACTCTGGCAGGCGCTGCCGGAAGAACTCCGCTTAAGTCCGCAT CGTTATCTGGCGACAAACAGTCCGCAGGGGCCGTGGTGGCTGCTCGGTTGGTGTGAGCGGGTGCCGGAAGCGGATGAGGTGCTGCCTGCGCCGCTGCCGCCGTACCGGGTACT GACCGGGCTGGTGGACCGCTTCGGGCGCACACAGACGTTCCACCGCGAAGCCGCCGGTGAATTCAGCGGCGAAATCACCGGCGTGACGGATGGTGCCTGGCGTCACTTCCGGC TGGTACTGACCACGCAGGCGCAGCGGGCAGAAGAAGCCCGGCAGCAGGCCATTTCCGGCGGGACGGAACCGTCCGCTTTTCCTGATACCCTGCCGGGTTACACCGAATATGGC CGGGACAACGGCATCCGTCTGTCTGCCGTGTGGCTGACGCACGACCCGGAATACCCGGAGAATTTACCTGCCGCGCCGCTGGTGCGCTATGGCTGGACGCCACGCGGCGAACT GGCGGTGGTGTATGACCGTAGTGGCAAACAGGTGCGCAGCTTTACTTACGATGATAAATACCGGGGCCGGATGGTGGCGCACCGTCACACGGGCCGGCCGGAAATCCGTTACC GTTACGACAGCGACGGGCGGGTGACAGAACAGCTAAACCCGGCAGGCTTAAGCTACACGTATCAGTATGAGAAAGACCGCATCACCATCACCGACAGCCTGGACCGCCGTGAA GTGCTGCACACGCAGGGCGAAGCCGGGCTGAAGCGGGTGGTGAAAAAGGAACACGCGGACGGCAGCGTCACGCAGAGTCAGTTTGACGCCGTGGGCAGGCTCAGGGCACAGAC GGATGCCGCAGGCAGGACAACAGAGTACAGCCCGGATGTGGTGACGGGCCTCATCACGCGCATAACCACGCCGGATGGCAGGGCATCGGCGTTTTACTATAACCACCACAACC AGTTAACGTCAGCCACCGGGCCTGACGGGCTGGAATTGCGCCGGGAATATGATGAATTGGGCCGTCTGATTCAGGAAACTGCCCCTGACGGCGATATCACCCGCTACCGTTAT GATAATCCACACAGTGACTTACCCTGCGCAACGGAAGATGCCACCGGCAGCCGGAAAACCATGACGTGGAGCCGTTACGGTCAGTTGCTGAGCTTCACCGACTGTTCCGGTTA TGTAACCCGTTATGACCATGACCGCTTCGGGCAGATGACGGCGGTGCACCGCGAGGAAGGGCTGAGTCAGTACCGCGCATACGACAGCCGTGGACAGTTAATTGCCGTGAAAG ACACGCAGGGCCATGAAACGCGGTATGAATACAACATCGCCGGTGACCTGACCGCCGTCATTGCCCCGGACGGCAGCAGAAACGGGACACAGTACGATGCGTGGGGAAAGGCC GTCCGTACCACGCAGGGCGGGCTGACGCGCAGTATGGAATACGATGCTGCCGGACGGGTCATCCGCCTGACCAGTGAAAACGGCAGCCACACCACCTTCCGTTACGATGTACT TGACCGGCTGATACAGGAAACCGGCTTTGACGGCCGCACACAGCGTTATCACCACGACCTGACCGGCAAACTTATCCGCAGCGAGGATGAGGGTCTGGTCACCCACTGGCACT ATGACGAAGCAGACCGCCTCACGCACCGCACCGTGAAGGGTGAAACCGCAGAGCGGTGGCAGTATGACGAACGTGGCTGGCTGACAGACATCAGCCATATCAGCGAAGGGCAC CGGGTGGCGGTGCATTACAGGTATGATGAGAAAGGCCGGCTGACCGGTGAGCGTCAGACGGTGCATCACCCGCAGACGGAAGCACTGCTCTGGCAGCATGAGACCAGACATGC GTACAACGCGCAGGGGCTGGCGAACCGCTGTATACCGGACAGCCTGCCCGCCGTGGAATGGCTGACCTACGGCAGCGGTTACCTGGCAGGCATGAAACTCGGCGACACACCGC TGGTGGAGTACACCCGCGACCGCCTGCACCGGGAAACGCTGCGCAGCTTCGGCCGTTATGAACTCACCACCGCTTATACCCCTGCCGGGCAGTTACAGAGCCAGCACCTGAAC AGCCTGCTGTCTGACCGCGATTACACCTGGAACGACAACGGCGAACTCATCCGCATCAGCAGCCCGCGCCAGACCCGGAGTTACAGCTACAGCACCACCGGCAGGCTGACCGG CGTTCACACCACCGCAGCGAATCTGGATATCCGCATCCCGTATGCCACAGACCCGGCAGGTAACCGCCTGCCCGACCCGGAGCTGCACCCGGACAGCACCCTCAGCATGTGGC CGGATAACCGTATCGCCCGTGACGCGCACTATCTTTACCGGTATGACCGTCACGGCAGGCTGACAGAGAAAACCGACCTCATCCCGGAAGGGGTTATCCGCACGGATGATGAG CGGACTCACCGGTACCATTACGACAGTCAGCACCGGCTGGTGCACTACACGCGGACACAATATGAAGAGCCGCTGGTCGAAAGTCGCTATCTTTACGACCCGCTGGGCCGCAG GGTGGCAAAACGGGTGTGGCGGCGTGAACGGGACCTGACGGGCTGGATGTCGCTGTCACGGAAACCGCAAGTGACCTGGTACGGCTGGGACGGCGACCGGCTGACCACGATAC AGAACGACAGGAGCCGCATCCAGACGATTTATCAGCCGGGGAGCTTCACGCCACTCATCAGGGTCGAAACTGCCACCGGTGAGCTGGCGAAAACGCAGCGCCGCAGCCTGGCG GATGCCCTTCAGCAGTCCGGCGGCGAAGACGGTGGCAGTGTGGTGTTCCCGCCGGTGCTGGTGCAGATGCTCGACCGGCTGGAAAGTGAAATCCTGGCTGACCGGGTGAGTGA GGAAAGCCGCCGCTGGCTGGCATCGTGCGGCCTGACCGTGGAGCAGATGCAAAACCAGATGGACCCGGTGTACACGCCGGCGCGAAAAATCCACCTGTACCACTGCGACCATC GCGGCCTGCCGCTGGCGCTCATCAGCACGGAAGGGGCAACAGCGTGGTGCGCAGAATATGATGAATGGGGCAACCTGCTGAATGAAGAGAACCCGCATCAGCTGCAGCAGCTT ATCCGGCTGCCGGGGCAGCAGTATGATGAGGAGTCCGGCCTGTATTACAACCGCCACCGCTATTATGACCCGCTGCAGGGGCGATATATCACTCAGGATCCGATTGGACTGAA GGGGGGATGGAACCTGTATGGATATCAATTGAATCCGATATCAGACATCGACCCCCTGGGTTTATCTATGTGGGAGGATGCAAAATCGGGGGCATGTACTAATGGTCTTTGCG GCACACTATCCGCTATGATAGGTCCAGATAAATTTGATTCTATAGATAGCACCGCATATGACGCCTTAAATAAAATAAATAGCCAATCTATTTGCGAAGATAAAGAGTTCGCT GGTTTAATATGTAAGGATAATAGTGGCAGATATTTCTCAACAGCACCTAACCGAGGAGAAAGAAAAGGATCATATCCATTCAATAGCCCTTGCCCTAATGGTACTGAGAAAGT ATCAGCTTATCATACTCATGGTGCAGATAGTCATGGAGAATATTGGGACGAAATATTTTCAGGTAAAGATGAGAAAATAGTTAAAAGTAAAGATAACAATATCAAGTCATTTT ATTTAGGTACGCCCAGTGGTAATTTTAAAGCAATAGATAACCACGGGAAGGAAATAACAAACAGAAAAGGATTACCTAATGTCTGCAGAGTTCATGGTAATATGTAA AAAAAATATTGTTTAGGAACTGTGTCATTGTATCTTTGTTTGTTTTTACA
first 50 base of yhhH gene
AATGTCTGCAGAGTTCATGGTAATATGTAAAAAAATATTGTTTAGGAACT
- Construct primers that have internal overlap with the resistance marker (pKD3/4) and external overlap with the target knockout gene.
- Forward primer: A
- CTTTTTCGTATGAAGATACTGTCATTAAAATAATAGAAAAGGATTTTACGGTGTAGGCTGGAGCTGCTTC
- Reverse primer: B
- AGTTCCTAAACAATATTTTTTTACATATTACCATGAACTCTGCAGACATTATGGGAATTAGCCATGGTCC
- Forward primer: A
- Construct primers that only flank the target gene for PCR verification
- Forward primer: C
- CTTTTTCGTATGAAGATACTGTCAT (25 bases)
- Reverse primer: D
- AGTTCCTAAACAATATTTTTTTACA (25 bases)
- Forward primer: C
pgl
- NC_000913 - pgl gene +50bp upstream +50bp downstream
GCGACTAATTTTAGCTGTTACAGTCAGTTGCTAAATGCAAAGGAGCATTCATGAAGCAAACAGTTTATATCGCCAGCCCTGAGAGCCAGCAAATTCACGTCTGGAATC TGAATCATGAAGGCGCACTGACGCTGACACAGGTTGTCGATGTGCCGGGGCAGGTGCAGCCGATGGTGGTCAGCCCGGACAAACGTTATCTCTATGTTGGTGTTCGCCCT GAGTTTCGCGTCCTGGCGTATCGTATCGCCCCGGACGATGGCGCACTGACCTTTGCCGCAGAGTCTGCGCTGCCGGGTAGTCCGACGCATATTTCCACCGATCACCAGGG GCAGTTTGTCTTTGTAGGTTCTTACAATGCGGGTAACGTGAGCGTAACGCGTCTGGAAGATGGCCTGCCAGTGGGCGTCGTCGATGTGGTCGAGGGGCTGGACGGTTGCC ATTCCGCCAATATCTCACCGGACAACCGTACGCTGTGGGTTCCGGCATTAAAGCAGGATCGCATTTGCCTGTTTACGGTCAGCGATGATGGTCATCTCGTGGCGCAGGAC CCTGCGGAAGTGACCACCGTTGAAGGGGCCGGCCCGCGTCATATGGTATTCCATCCAAACGAACAATATGCGTATTGCGTCAATGAGTTAAACAGCTCAGTGGATGTCTGG GAACTGAAAGATCCGCACGGTAATATCGAATGTGTCCAGACGCTGGATATGATGCCGGAAAACTTCTCCGACACCCGTTGGGCGGCTGATATTCATATCACCCCGGATGG TCGCCATTTATACGCCTGCGACCGTACCGCCAGCCTGATTACCGTTTTCAGCGTTTCGGAAGATGGCAGCGTGTTGAGTAAAGAAGGCTTCCAGCCAACGGAAACCCAGC CGCGCGGCTTCAATGTTGATCACAGCGGCAAGTATCTGATTGCCGCCGGGCAAAAATCTCACCACATCTCGGTATACGAAATTGTTGGCGAGCAGGGGCTACTGCATGAA AAAGGCCGCTATGCGGTCGGGCAGGGACCAATGTGGGTGGTGGTTAACGCACACTAACCGCTGATTTACCCGGCGCAGTCTCTCCTGCGCCGGTGTATTAACCTATC
- Construct primers that have internal overlap with the resistance marker (pKD3/4) and external overlap with the target knockout gene.
- Forward primer: A
- GCGACTAATTTTAGCTGTTACAGTCAGTTGCTAAATGCAAAGGAGCATTCGTGTAGGCTGGAGCTGCTTC
- Reverse primer: B
- GATAGGTTAATACACCGGCGCAGGAGAGACTGCGCCGGGTAAATCAGCGGATGGGAATTAGCCATGGTCC
- Forward primer: A
- Construct primers that only flank the target gene for PCR verification
- Forward primer: C
- GCGACTAATTTTAGCTGTTACAGTC (25 bases)
- Reverse primer: D
- GATAGGTTAATACACCGGCGCAGGA (25 bases)
- Forward primer: C
ptsG
- NC_000913 - ptsG gene +50bp upstream +50bp downstream
CACGCGTGAGAACGTAAAAAAAGCACCCATACTCAGGAGCACTCTCAATTATGTTTAAGAATGCATTTGCTAACCTGCAAAAGGTCGGTAAATCGCTG ATGCTGCCGGTATCCGTACTGCCTATCGCAGGTATTCTGCTGGGCGTCGGTTCCGCGAATTTCAGCTGGCTGCCCGCCGTTGTATCGCATGTTATGGCAG AAGCAGGCGGTTCCGTCTTTGCAAACATGCCACTGATTTTTGCGATCGGTGTCGCCCTCGGCTTTACCAATAACGATGGCGTATCCGCGCTGGCCGCAGT TGTTGCCTATGGCATCATGGTTAAAACCATGGCCGTGGTTGCGCCACTGGTACTGCATTTACCTGCTGAAGAAATCGCCTCTAAACACCTGGCGGATACT GGCGTACTCGGAGGGATTATCTCCGGTGCGATCGCAGCGTACATGTTTAACCGTTTCTACCGTATTAAGCTGCCTGAGTATCTTGGCTTCTTTGCCGGTA AACGCTTTGTGCCGATCATTTCTGGCCTGGCTGCCATCTTTACTGGCGTTGTGCTGTCCTTCATTTGGCCGCCGATTGGTTCTGCAATCCAGACCTTCTC TCAGTGGGCTGCTTACCAGAACCCGGTAGTTGCGTTTGGCATTTACGGTTTCATCGAACGTTGCCTGGTACCGTTTGGTCTGCACCACATCTGGAACGTA CCTTTCCAGATGCAGATTGGTGAATACACCAACGCAGCAGGTCAGGTTTTCCACGGCGACATTCCGCGTTATATGGCGGGTGACCCGACTGCGGGTAAAC TGTCTGGTGGCTTCCTGTTCAAAATGTACGGTCTGCCAGCTGCCGCAATTGCTATCTGGCACTCTGCTAAACCAGAAAACCGCGCGAAAGTGGGCGGTAT TATGATCTCCGCGGCGCTGACCTCGTTCCTGACCGGTATCACCGAGCCGATCGAGTTCTCCTTCATGTTCGTTGCGCCGATCCTGTACATCATCCACGCG ATTCTGGCAGGCCTGGCATTCCCAATCTGTATTCTTCTGGGGATGCGTGACGGTACGTCGTTCTCGCACGGTCTGATCGACTTCATCGTTCTGTCTGGTA ACAGCAGCAAACTGTGGCTGTTCCCGATCGTCGGTATCGGTTATGCGATTGTTTACTACACCATCTTCCGCGTGCTGATTAAAGCACTGGATCTGAAAAC GCCGGGTCGTGAAGACGCGACTGAAGATGCAAAAGCGACAGGTACCAGCGAAATGGCACCGGCTCTGGTTGCTGCATTTGGTGGTAAAGAAAACATTACT AACCTCGACGCATGTATTACCCGTCTGCGCGTCAGCGTTGCTGATGTGTCTAAAGTGGATCAGGCCGGCCTGAAGAAACTGGGCGCAGCGGGCGTAGTGG TTGCTGGTTCTGGTGTTCAGGCGATTTTCGGTACTAAATCCGATAACCTGAAAACCGAGATGGATGAGTACATCCGTAACCACTAA TCCGTAAGACGTTGGGGAGACTAAGGCAGCCAGATGGCTGCCTTTTTTAC
- Construct primers that have internal overlap with the resistance marker (pKD3/4) and external overlap with the target knockout gene.
- Forward primer: A
- CACGCGTGAGAACGTAAAAAAAGCACCCATACTCAGGAGCACTCTCAATTGTGTAGGCTGGAGCTGCTTC
- Reverse primer: B
- GTAAAAAAGGCAGCCATCTGGCTGCCTTAGTCTCCCCAACGTCTTACGGAATGGGAATTAGCCATGGTCC
- Forward primer: A
- Construct primers that only flank the target gene for PCR verification
- Forward primer: C
- CACGCGTGAGAACGTAAAAAAAGCA (25 bases)
- Reverse primer: D
- GTAAAAAAGGCAGCCATCTGGCTGC (25 bases)
- Forward primer: C
Experiment
- We will use this protocol Single-gene knockouts using λ red system
*It was a bad idea we have not enough time !
SID
Colicin is a type of bacteriocin produced by and toxic to some strains of Escherichia coli. Colicins are released into the environment to reduce competition from other bacterial strains. Colicins bind to outer membrane receptors, using them to translocate to the cytoplasm or cytoplasmic membrane, where they exert their cytotoxic effect, including depolarisation of the cytoplasmic membrane, DNase activity, RNase activity, or inhibition of murein synthesis.
We are interested in two of them :
- Colicin E2 (DNase activity, Tol-dependent)
- Colicin D (RNase activity, TonB-dependent)