IGEM:IMPERIAL/2006/project/popsblocker/Implementation: Difference between revisions
From OpenWetWare
Jump to navigationJump to search
No edit summary |
No edit summary |
||
| Line 11: | Line 11: | ||
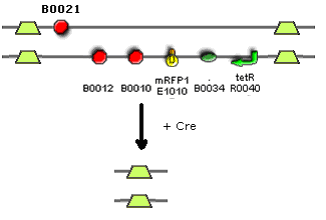
[[Image:Cre.PNG]] | [[Image:Cre.PNG]] | ||
Stop Codons | |||
I Have included a stop codon between lox66 and B0021 to prevent the e-coli cells from translating past the first lox site. This reduces the load that this part will place on the E.coli cell so increases it’s stability in vivo. I checked the E-coli codon usage database on NCBI and found that TGA was by far the most commonly used in nature. | |||
Frameshift | |||
The Lox sites are 34bp long therefore if this part is placed after an RBS it will induce a frameshift in the protein produced, therefore creating a mangled protein. I added two random bases to the start of the lox site to prevent a frameshift. | |||
==Ligation strategy== | ==Ligation strategy== | ||
Revision as of 13:29, 29 October 2006
Stop Codons
I Have included a stop codon between lox66 and B0021 to prevent the e-coli cells from translating past the first lox site. This reduces the load that this part will place on the E.coli cell so increases it’s stability in vivo. I checked the E-coli codon usage database on NCBI and found that TGA was by far the most commonly used in nature.
Frameshift
The Lox sites are 34bp long therefore if this part is placed after an RBS it will induce a frameshift in the protein produced, therefore creating a mangled protein. I added two random bases to the start of the lox site to prevent a frameshift.
Ligation strategy
- Apart from ligating into the necessary plasmid at the end, there are no ligation steps involved in making this device. PCR and PCR fusion are being are used because of the unusual design of the device, in so far as both strands have coding regions. The link below explains the method by which we will use PCR to create the device.
Primers
Design of primers and method of construction
Commercially available Cre plasmid
WetLab work entries
Jonny Wells <br\> Kirsten Jensen
(08/06 - 10/06)