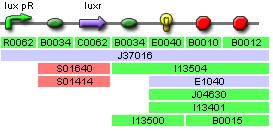
IGEM:IMPERIAL/2006/project/parts/BBa J37016
This is a document for part BBa_J37016 AND BBa_J37020!!! (they are the same part where one produces polycistronic RNA and the other doesn't
SPECIFICATIONS
Design a part to
- To measure the rate of AHL-lactonase synthesis as a function of the amount of AHL present in the medium
- To measure the Km and Vm of LuxR in our cells.
Background
This expeiment will characterise the transfer function of the parts J37016 and J37020. One of these parts will form the reciver in the predator cell. There are many similarities between this characterisation and that of the prey reciver
Design
These two parts are very similar and have the same inputs and outputs, therefore they can be analysed in exactly the same way
These designs should meet the specifications mentioned above and operate as shown in fig1
The differences are in the rates of production of GFP to LuxR. Part J37016 will produce a polycistronic strand of mRNA and the rate of GFP to LuxR translation will depend on which RBS is farthest upstream. The second design; Part J37020 produces two strands of mRNA they are produced by the same promoter, LuxpR but the seuqance surrounding luxpR differs between the two sites so there may be differing rates of transcription between the promoters controlling GFP and luxR. A more reliable method to get the luxR rates of expression would be to tag LuxR with an antigen then run an assay for it. This would however require a very large amount o lab work and the modls do not need luxR translation, simply the transfer function of HSL -> aiiA
MODELLING
- Input = AHL
- Output = GFP
The model produces GFP and LuxR as a function of LuxR + AHL
An interesting dynamic has been observed in this system.
- The rate of synthesis of luxR depends on the AHL concentration and the luxR concentration and a constant
- The rate of degredation of LuxR depends on only the LuxR constant and the rate of degredation
This means that the rate of degredation can be higher than the rate of syntheses so the steady state will be no LuxR for very low values of AHL but after a critical value of AHL the steady state will be positive.

Values (unrealistic)
Unrealistic Values Name Value LuxR 0.8 AHL (Low) 0.07 AHL (High) 0.11 GFP 0
Parameters (Un-realistic)
Unrealistic Perameters Name Value Vm LuxR 2 Km LuxR 1.87 K_Deg_LuxR 0.1 K_Deg_GFP 0.1
- We assume HSL is constant
- (the gene expression was moddeled using mechiles menten kenitics so luxR has a Km and Vm of binding to the gene)
Key paremeter - Km of LuxR
This must be high. We can increace the apparant Km in real cells using recombinant plasmids. A high Km will reduce the rate of reaction and increace the range at which AHL can alter the reaction.
To work out Km and Vm
Transcription at lux pR is proportional to the amount of LuxR+HSL present. This is dependent on the amount of HSL added so the rate of GFP production will be proportional to the amount of HSL added.
If we know the Rate of GFP degradation and the equilibrium conc of GFP then we can work out the rate of GFP synthesis for that amount of HSL as degradation = synthesis at equilibrium.
If we know the rates and the substrate conc for those rates we can make a lineweaver burke plot. Plot 1/v against 1/[s].
1 / V = (Km / Vm)(1 / [S]) + (1 / Vm)
Y = M X + C
[:http://openwetware.org/images/3/3b/Lineweaver_Buke_Plot.JPG]
This will allow us to work out the real values of Km and Vm for LuxR+AHL when it binds LuxPr, :-D
Transfer Function
The transfer function of this part shows an unusual property in that the system will not produce any GFP at low HSL values (rather than tending to 1/infinity). This is caused by the critical value of the system described above. This should make this device function as a low pass filter, this property is of no use to the oscilator, but should be noted. The predator cells will not become de-sensitised to HSL because the luxpR operon shows leaky expression, this keeps a low conc of LuxR in the cells at all times (this property was not modeled).
Implementation
These two parts are very similar and have the same inputs and outputs, therefore they can be analysed in exactly the same way
This construct is used to characterise the pLuxR transfer function, and noteably its effect on YFP expression, which would be equivalent to AiiA expression in the full system with one promoter.
This construct is used to characterise the pLuxR transfer function, and noteably its effect on YFP expression, which would be equivalent to AiiA expression in the full system with two promoters.
The input is HSL and the outputs are LuxR and GFP. GFP will be mesured
VALIDATION / TESTING
- Inoculate a culture from a single bacterial colony in LB growth medium containing appropriate antibiotic.
- Incubate at 37oC for 15 hours in a shaker.
- Remove cultures from shaker and dilute by a factor of 10, by adding 1ml of culture into 9ml of fresh LB medium.
- Incubate at 37oC for 2 hours in a shaker - This returns cells to exponential phase
- We have a stock AHL solution of 1000uM
- Serial dilute to give final 1ml stock concentrations of 1000uM, 100uM, 10uM, 1uM, 100nM, 10nM, 1nM (check the -20oC Freezer as these may be already made up).
- Add the following to eight different labelled ependorf tubes (remember to add the AHL last):
| Sample (ul) | Stock Concentration | AHL (ul) | Final AHL Concentration |
|---|---|---|---|
| 990 | 1000uM | 10 | 10uM |
| 990 | 100uM | 10 | 1uM |
| 990 | 10uM | 10 | 100nM |
| 990 | 1uM | 10 | 10nM |
| 990 | 100nM | 10 | 1nM |
| 990 | 10nM | 10 | 0.1nM |
| 990 | 1nM | 10 | 0.01nM |
| 1000 | N/A | 0 | 0nM |
- Add 200uL of growth medium to a well to act as a control
- Vortex tubes for 1 second
- Incubate the culture in a 37oC incubator, taking readings every 30minutes
- Taking readings
- Prepare an ice bucket containing a pre-chilled 96 well plate - The ice is used to stop cell activity, therefore its imporant the plate stays cold
- Remove the tubes from the incubator
- Pippet 200ul samples from each tube into the 96 well plate inside the ice bucket
- Vortex the tubes for 1 second - As the cells aren't in the shaker, the vortex helps to airate the culture
- Return tubes to the oven
- Repeat for a total of 4 measurments
- Take 96 well plate in ice bucket to the SAF
- Record data from the fluorimeter
References
Tom Hinson
Comments
Tom:
- Do we need J37020??
- The only difference is having an extra promoter
- This can be achieved by ligating a promoter onto the end of J37016
- Although this will take one more ligation, we don't really know how useful having two promoters will be
- IE I think it will be better to get data from J37016 before deciding to add an extra promoter
- How do you measure LuxR Km and Vm (In specifications)
- LuxR is varying in response to AHL
- We have no way of measuring LuxR expression
Christin:
- If we want to add a single promoter at the end of J37016, we would need 5 ligation steps, whereas ligating parts J37016 and J37020 at the same time would take 4 steps each
- We could build J37020 at the same time and then test the two parts together, thus saving time to build it afterwards if we decide we want to use this part later after all (for whatever reason)
- If we are absolutely sure about using J37016 though and do not need J37020, I agree with Tom's comment above
Resolution (29 July)
- We will be making both parts and testing them to see which one is more effective
- RESOLVED ALL ISSUES