IGEM:Harvard/2006/Transcription vs Protein circuits
It is pretty easy to see that the following circuit will produce an oscillator. The forward "activation" arrows establish a net forward movement, and the back "repression" arrows prevent the circuit from reaching steady state with all elements "on".

But what do these arrows actually mean? One could imagine that the arrows represent transcription factor activity. Each circle represents an operon that produces proteins that repress or promote transcription of other operons.
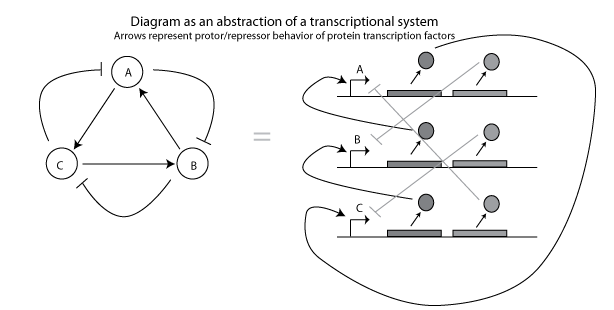
This is the basis for synthetic circuits such as the Ellowitz repressilator. However, it appears that many biological oscillators, such as the KaiA/B/C system of cyanobacteria, do not create oscillations merely from transcriptional dynamics.
Instead, transcription remains constant and it is the catalytic behavior of proteins that oscillates, based on post-translational modifications such as phosphorylation or methylation.
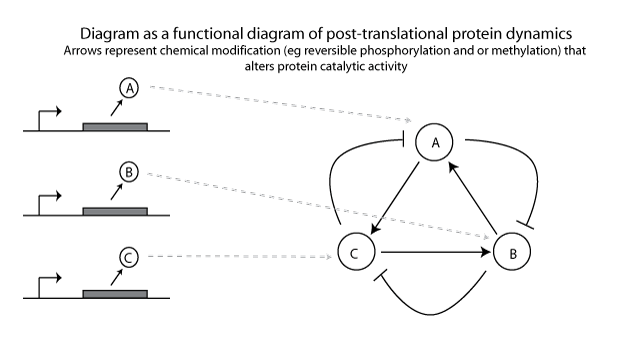
As you can see, the methods required to construct the two different implementations of the same abstract system are very different. The functional constraints are very different as well; the dynamics of the transcription-based circuit, for example, will be limited by the upper-limit of transcription/translation speeds. The post-translation-based circuit will depend more on the reaction rates for phosphorylation/dephosphorylation.