Drummond:Research: Difference between revisions
Dadrummond (talk | contribs) |
Dadrummond (talk | contribs) |
||
| Line 8: | Line 8: | ||
We are pursuing mechanistic answers to these questions, taking a biochemical and genetic approach, with an emphasis on developing high-resolution, high-mass-accuracy mass spectrometric techniques for proteome-scale quantitation. From a theoretical standpoint, we are interested in understanding the imprints that natural selection on fidelity and misfolding leave on evolving genes and genomes. | We are pursuing mechanistic answers to these questions, taking a biochemical and genetic approach, with an emphasis on developing high-resolution, high-mass-accuracy mass spectrometric techniques for proteome-scale quantitation. From a theoretical standpoint, we are interested in understanding the imprints that natural selection on fidelity and misfolding leave on evolving genes and genomes. | ||
<hr/> | <hr/> | ||
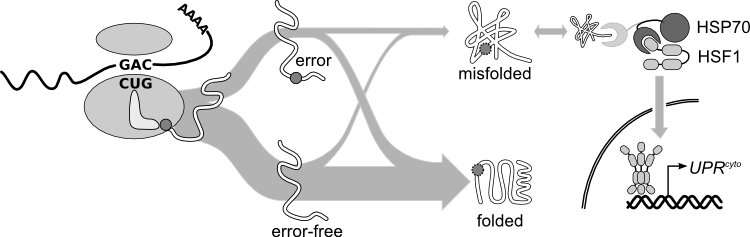
==Spectrum and | ==Spectrum, frequency, and consequences of errors in protein synthesis== | ||
At the canonical translational misreading error rate of 1 error per 2000 amino acids translated, <em>one in five</em> average-length proteins will contain at least one amino acid not encoded in the messenger RNA. Single amino-acid substitutions frequently destabilize proteins, sometimes disrupting protein folding. Historical measurements of translational error focus on single codons, often in heterologous reporter constructs, leaving open basic questions, such as how the frequency of translation errors varies between proteins. | At the canonical translational misreading error rate of 1 error per 2000 amino acids translated, <em>one in five</em> average-length proteins will contain at least one amino acid not encoded in the messenger RNA. Single amino-acid substitutions frequently destabilize proteins, sometimes disrupting protein folding. Historical measurements of translational error focus on single codons, often in heterologous reporter constructs, leaving open basic questions, such as how the frequency of translation errors varies between proteins. | ||
Revision as of 17:29, 15 February 2012
What are the spectrum, frequency and consequences of errors in protein synthesis? How do cells sense and respond to misfolded proteins, particularly in the eukaryotic cytosol? How does stochasticity in protein synthesis alter the composition and stability of the proteome? What benefits do errors confer? Does error-induced protein misfolding influence the progression of neurodegenerative diseases such as ALS?
We are pursuing mechanistic answers to these questions, taking a biochemical and genetic approach, with an emphasis on developing high-resolution, high-mass-accuracy mass spectrometric techniques for proteome-scale quantitation. From a theoretical standpoint, we are interested in understanding the imprints that natural selection on fidelity and misfolding leave on evolving genes and genomes.
Spectrum, frequency, and consequences of errors in protein synthesis
At the canonical translational misreading error rate of 1 error per 2000 amino acids translated, one in five average-length proteins will contain at least one amino acid not encoded in the messenger RNA. Single amino-acid substitutions frequently destabilize proteins, sometimes disrupting protein folding. Historical measurements of translational error focus on single codons, often in heterologous reporter constructs, leaving open basic questions, such as how the frequency of translation errors varies between proteins.
For the past seven years, we have assembled evidence that high-expression proteins are under selection to reduce errors (increased translational accuracy), and to fold properly despite the errors that do occur (increased translational robustness). We are using high-resolution, high-mass-accuracy mass spectrometry to survey the actual sequences of proteins in vivo, and to address these basic questions about variability in error rates and consequences across the proteome.
Consequences of synthesis errors
Response to misfolded proteins
Under construction...thank you for your patience as we remodel to give you greater insight, improved clarity, and more images.