DNAmazingMission: Difference between revisions
| Line 78: | Line 78: | ||
=Motivation= | =Motivation= | ||
[[Image:compare.png|500px|right]] | [[Image:compare.png|500px|right]] | ||
At the beginning of this project, we planned to synthesize a complicated "city-traffic" like DNA Origami system on which several DNA motors operate. During the planning, we recognize that there were slots in the current CAD programs which may be needed to fill in: the "one-click" design for complicated structures and the addition of sticky ends. The current approach in CAD of DNA Origami requires the users manually design the scaffold way and select the position of cross-overs which may be tedious and may impossible for very complex structures in the future. Furthermore, sticky ends have not been well-considered. Thus, we decided to start at developing a suitable CAD program for our purposes like Rothemund did in 3 months before he published the first paper about DNA Origami in 2006. We admitted this was a challenge as we had a very different approach to the algorithm of the CAD program, and there have been very few well-documented available instructions in DNA Origami CAD programs. Furthermore, none of us has been trained in the computing science. However, recognizing the benefits of our movement, we insisted to carry on with the project. We believe this would be a good start for our experimental projects in the future as well as a good contribution to the community of DNA Origami. | <p align="left"> | ||
At the beginning of this project, we planned to synthesize a complicated "city-traffic" like DNA Origami system on which several DNA motors operate. During the planning, we recognize that there were slots in the current CAD programs which may be needed to fill in: the "one-click" design for complicated structures and the addition of sticky ends. The current approach in CAD of DNA Origami requires the users manually design the scaffold way and select the position of cross-overs which may be tedious and may impossible for very complex structures in the future. Furthermore, sticky ends have not been well-considered. Thus, we decided to start at developing a suitable CAD program for our purposes like Rothemund did in 3 months before he published the first paper about DNA Origami in 2006. We admitted this was a challenge as we had a very different approach to the algorithm of the CAD program, and there have been very few well-documented available instructions in DNA Origami CAD programs. Furthermore, none of us has been trained in the computing science. However, recognizing the benefits of our movement, we insisted to carry on with the project. We believe this would be a good start for our experimental projects in the future as well as a good contribution to the community of DNA Origami.</p> | |||
=Objectives= | =Objectives= | ||
Latest revision as of 23:04, 2 November 2011
<html><head> <style type="text/css"> .main { margin: 0 auto; width: 804px;border-style:solid; border-width:5px; }
- logo{ margin:10px 0 0 52px; display:block; background:url('http://openwetware.org/images/7/74/Banner20.png') 0 0 no-repeat; width:700px; height:200px; text-indent:-9999px;}
.navbox { position: relative; float: left; }
- menu{ background:url('http://openwetware.org/images/3/3f/Menu_bg.png') 0 0 no-repeat; width:643px; height:100px; margin:51px 0 0 78px; padding-top:100px}
- menu li{ float:left;}
- menu a{ font-size:30px; color:#000000; line-height:1.2em; text-decoration:none; letter-spacing:-1px;}
.nav1{ padding:26px 0 0 37px;} .nav2{ padding:16px 0 0 30px;} .nav3{ padding:36px 0 0 17px;} .nav4{ padding:16px 0 0 19px;} .nav5{ padding:26px 0 0 30px;}
- menu .nav1 a:hover{ color:#bb0e0e}
- menu .nav2 a:hover{ color:#ca6509}
- menu .nav3 a:hover{ color:#3f9711}
- menu .nav4 a:hover{ color:#0ca0ce}
- menu .nav5 a:hover{ color:#8606c5}
</style> </head>
<div class="extra"> <div id="page_1"> <div class="main" style="padding-top:10px; padding-bottom:10px"> <!--header --> <header> <a href="index.html" id="logo">DNAmazing asfsaf. Smart. Effective</a> <nav> <ul id="menu"> <li class="nav1"><a href="http://openwetware.org/wiki/User:NUS_Dnamazing">Home</a></li> <li class="nav2"><a href="http://openwetware.org/wiki/DNAmazingMission">Mission</a></li> <li class="nav3"><a href="http://openwetware.org/wiki/DNAmazingProcess">Process</a></li> <li class="nav4"><a href="http://openwetware.org/wiki/DNAmazingResult">Results</a></li> <li class="nav5"><a href="http://openwetware.org/wiki/DNAmazingResources">Resources</a></li> </ul> </nav> </header> <!--header end-->
</div>
</div>
</div>
</html>
Inspiration

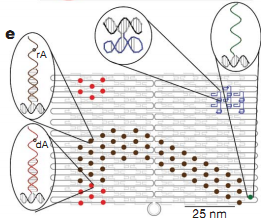

Since the first paper published by Paul W.K.Rothemund about DNA Origami in 2006, there have been intense efforts observed in the community of nanoscientists, chemists, and nanoengineers to push the field ahead. During only 5 years, DNA Origami has advanced from 2D to 3D nanostructures, from DNA Origami systems in relaxing state to systems intentionally in tension to create curves, from simple structures to sophisticate structures. DNA Origami is definitely one of most promising tools for nanoengineering today.
With its more than tenfold higher resolution than that of any previous techniques and its capability to create complex nanostructures from bottom up, we believe DNA Origami can be employed as the molds or platforms with connection sites at its sticky ends for other nano-devices and components. To illuminate, The rigidity of DNA Origami structures together with the stability of their sticky ends can be used to create the platforms on which DNA nanomachine can operate by hoping around sticky ends sites. We strongly believe that in the near future we may be able to construct nano-cities in which nano-motocycles go around and transport molecules to nano-factories. In fact, there have been few initial efforts to realize this vision from Seeman's group, Erik Winfree's group,Hao Yan's group and Pierce's group (Figure 1 and Figure 2).
DNA Origami has been proved to provide access to the synthesis of any desired shape. This ability strongly attracts scientists in the field of nanoengineering who are finding ways to fabricate complicated nanostructures for biomedical and nanoelectronic applications. The top-down approach is struggling with the problems of resolution and environmental issues; the bottom-up approach in colloid science is facing the problems of uniform sizes and kinetic as well as thermodynamic limitations. The emerging DNA Origami technique seems to be a new candidate to handle these difficulties.It has been reported some promising developments in the field of biomineralization which can transform peptide and DNA structures into metallic system. Toether with biomineralization, DNA Origami can be employed in the field of nanofrabrication. In the near future, a blueprint of a complicated nanocircuit will be fabricated in the form of DNA Origami structures which will be converted to the metallic circuit by biomineralization. This requires a new CAD program which is able to provide the users high level of complexity in the structures and "one-click" to the result to accelerate the processing time of design in industry.
The researching thrust in DNA Origami is no doubt turning to the applications of this technique from the fabrication of various shapes which has been well established; we believe complexity and the addition of stick ends will be the most important factors to advance the field in the future.
Motivation

At the beginning of this project, we planned to synthesize a complicated "city-traffic" like DNA Origami system on which several DNA motors operate. During the planning, we recognize that there were slots in the current CAD programs which may be needed to fill in: the "one-click" design for complicated structures and the addition of sticky ends. The current approach in CAD of DNA Origami requires the users manually design the scaffold way and select the position of cross-overs which may be tedious and may impossible for very complex structures in the future. Furthermore, sticky ends have not been well-considered. Thus, we decided to start at developing a suitable CAD program for our purposes like Rothemund did in 3 months before he published the first paper about DNA Origami in 2006. We admitted this was a challenge as we had a very different approach to the algorithm of the CAD program, and there have been very few well-documented available instructions in DNA Origami CAD programs. Furthermore, none of us has been trained in the computing science. However, recognizing the benefits of our movement, we insisted to carry on with the project. We believe this would be a good start for our experimental projects in the future as well as a good contribution to the community of DNA Origami.
Objectives
As the first version of DNAmazing is a student project expected to lay out the foundation for the next generation of CAD program and 3 months are not long enough to perfect the project, we planned to provide the users with basic functions and introduction to the new approach to CAD programs in DNA Origami:
1. "One-click to results" feature which includes:
- Automatically generate folding path
- Automatically determine crossover position
- Automatically generate DNA staple sequence
- Automatically merge the staple fragments
2. Addition of sticky ends.
3. Thermodynamic calculation to determine the thermal stability of stick ends
Vision
To the best of our knowledge, DNAmazing is the only student project to develop a CAD program in DNA Origami from the scratch. As being developed in only 3 months, DNAmazing could not function perfectly like other professional software. However, we believe DNAmazing is a success project for us. We decided to put DNAmazing on Open Source so that people around the world, including students like us,can refer to when developing a new CAD software, contributing to DNAmazing or even just having fun. DNAmazing was written in popular C# language on Visual Studio 2010 IDE without using any third parties library and was simple enough for the others to understand and develop the algorithm. There are many potential issues which DNAmazing is facing: the long processing time for large structures, better GUI interface, extension from 2D structures to 3D structures. Personally, we may develop the program more after Biomod so that we can use it to conduct new lab experiments. We hope that DNAmazing will bring a fresh breath to the field and perhaps envisage the new path for the development of the CAD programs.