Courtney L. Merriam Week 11
Purpose
This journal’s purpose is to work towards the completion of the weekly assignment for week eleven. We are examining amino acid sequences and comparing them to find a region that displays a significant amount of genetic difference.
Methods and Results
Work Session for Our Project
- Reselect the subjects for out experiment
- Subjects 13,12,10, and 4 the first and the last visit. Subjects 4 and 10 are the highest progressing subjects and have based on their annual CD4 count. Subject 12 and 13 were the lowest non-progressing subjects based on their annual CD4 Count
- Performed a multiple sequences alignment with all subjects chosen and their clones from first and last visits
- Analyzed data to find a pattern in the amino acid changes
- The Huang paper sequence was compare to the Markham sequences using a protein rendering and analysis program
- The purpose behind this was to find the differences between the Huang paper and the Markham sequences
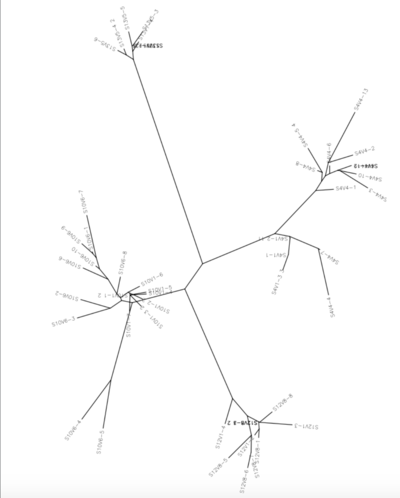
- Perform a cluster alignment of all the sequences we are using to show the difference between each subject's first and last visit
- The distances were recorded for each subject.
- Create a presentation of the work in progress for next week
Data and Files

- This structure was used to generate the Markham sequences to be compared to experimental subject sequences for comparison.
ClustalDist Results
- Following data is the distance between each subject's first and last visit
- Subject 10: 0.116
- Subject 4: 0.117
- Subject 12: 0.053
- Subject 13: 0.021
Experimental Subjects
Rapid Progressors
- Subject 13-visits 1 and 5
- Subject 12-visits 1 and 8
Non Progressors
- Subject 10-vists 1 and 6
- Subject 4-visits 1 and 4
Amino Acid Mutations Between Subject's First and Last Visits
- Subject 4→ 1 mutation
- Subject 10→ 8 mutations
- Subject 12→ 3 mutations
- Subject 13→ 0 mutations
Conclusion
Continuing the same series of experiments, we used the data we acquired in the previous week. The various progressors amino acid mutations were analyzed witha multicple sequences alignment, which did not show major differences between the several subjects first and last visits. An analysis from ClustalDist also showed that not all subjects had noticeable change in amino acid, though some did. Using a 3d modelling tool, we mapped an amino acid sequences to find the specific area that was commonly mutated. More than fifty percent of each subjects mutations occurred between amino acids 38 and 42 out of a 95 amino acid long sequence on the V3 region.
Acknowledgments
I collaborated with Shivum A Desai in class on this assignment. While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source.
- Courtney L. Merriam 16:36, 8 November 2016 (EST):
References
- Kirchherr, J. L., Hamilton, J., Lu, X., Gnanakaran, S., Muldoon, M., Daniels, M., Kasongo, W., Chalwe, V., Mulenga, C., Mwananyanda, L., Musonda, R.M., Yuan, X., Montefiori, D.C., Korber, M.T., Haynes, B.F., & Musonda, R. M. (2011). Identification of amino acid substitutions associated with neutralization phenotype in the human immunodeficiency virus type-1 subtype C gp120. Virology, 409(2), 163-174. DOI: 10.1016/j.virol.2010.09.031
- Markham, R.B., Wang, W.C., Weinstein, A.E., Wang, Z., Munoz, A., Templeton, A., Margolick, J., Vlahov, D., Quinn, T., Farzadegan, H., & Yu, X.F. (1998). Patterns of HIV-1 evolution in individuals with differing rates of CD4 T cell decline. Proc Natl Acad Sci U S A. 95, 12568-12573. dos: 10.1073/pnas.95.21.12568
Useful Links
Clas Page: Bioinformatics Laboratory
| Weekly Assignments | Individual Journal Assignments | Shared Journal Assignments |
|---|---|---|
|
|
|