CH391L/S2013 Taylor Pursell Feb 13 2013: Difference between revisions
| (One intermediate revision by the same user not shown) | |||
| Line 51: | Line 51: | ||
They then tested various conditions to see their effect on the nucleation and growth rates. First, they modulated the concentration of magnesium acetate ([Me(Oac)2), which associates with ATP. As the concentration of Me(Oac)2 was increased the rate of nucleation increased, but it had no effect on growth rate suggesting that the presence of Mg+-ATP cofactor is critical for increasing the rate of nucleation of RecA, but not in growth rate. The effects of ATP hydrolysis was tested using ATP, ATP-γ-S (an non-hydrolyzable analog), and dATP (a more readily hydrolysable version of ATP). In the presence of ATP-γ-S and dATP, the nucleation rate is 10-fold and 5-fold faster, respectively, than with ATP, suggesting that hydrolysis has no bearing on the stability of the filament. The growth rate was not statistically effected by ATP-γ-S or dATP compared to ATP. These results suggest that ATP acts as a conformational regulator, increasing RecA protein’s affinity for ssDNA upon binding. | They then tested various conditions to see their effect on the nucleation and growth rates. First, they modulated the concentration of magnesium acetate ([Me(Oac)2), which associates with ATP. As the concentration of Me(Oac)2 was increased the rate of nucleation increased, but it had no effect on growth rate suggesting that the presence of Mg+-ATP cofactor is critical for increasing the rate of nucleation of RecA, but not in growth rate. The effects of ATP hydrolysis was tested using ATP, ATP-γ-S (an non-hydrolyzable analog), and dATP (a more readily hydrolysable version of ATP). In the presence of ATP-γ-S and dATP, the nucleation rate is 10-fold and 5-fold faster, respectively, than with ATP, suggesting that hydrolysis has no bearing on the stability of the filament. The growth rate was not statistically effected by ATP-γ-S or dATP compared to ATP. These results suggest that ATP acts as a conformational regulator, increasing RecA protein’s affinity for ssDNA upon binding. | ||
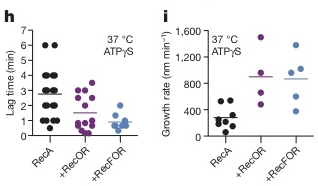
Next, the effect of mediator proteins were tested, specifically RecO, RecR, and RecF, genetically and biochemically established mediator proteins in E. coli [1]. They tested two combinations: the RecOR complex which interacts with SSB to mediate assembly in a structure-independent manner, and RecFOR complex which acts at the junction of ssDNA and dsDNA [1]. To test the effect of these cofactors, a new set up was used described in the Methods section above. In the presence of RecOR, the mean lag time decreased, some occurring in less time than RecA alone. The addition of RecF further decreased the mean lag time (Figure 4h). RecOR accelerated the growth rate but RecF had no effect (Figure 4i). | Next, the effect of mediator proteins were tested, specifically RecO, RecR, and RecF, genetically and biochemically established mediator proteins in E. coli [1]. They tested two combinations: the RecOR complex which interacts with SSB to mediate assembly in a structure-independent manner, and RecFOR complex which acts at the junction of ssDNA and dsDNA [1]. To test the effect of these cofactors, a new set up was used described in the Methods section above. In the presence of RecOR, the mean lag time decreased, some occurring in less time than RecA alone. The addition of RecF further decreased the mean lag time (Figure 4h, below). RecOR accelerated the growth rate but RecF had no effect (Figure 4i, below). | ||
[[Image: | [[Image: Fig4_HI.png]] | ||
Based on these results, the researches created a model (Figure 4j) in which RecOR interacts with SSB to facilitate RecA assembly [1,7] by trapping transiently unbound ssDNA thus weakening the SSB-ssDNA binding and facilitating RecA binding [1]. | Based on these results, the researches created a model (Figure 4j, below) in which RecOR interacts with SSB to facilitate RecA assembly [1,7] by trapping transiently unbound ssDNA thus weakening the SSB-ssDNA binding and facilitating RecA binding [1]. | ||
[[Image: Fig4_ProposedModel.png]] | |||
==Significance/Applications== | ==Significance/Applications== | ||
Latest revision as of 10:04, 11 February 2013
“Direct imaging of RecA nucleation and growth on single molecules of SSB-coated ssDNA” Jason C. Bell, Jody L. Plank, Christopher C. Dombrowski, and Stephen C. Kowalczykowski
Background
To ensure their survival, cells have evolved mechanisms by which they both monitor genome integrity and repair damage. The most dangerous type of DNA damage is double stranded breaks (DSB) which can be caused by endogenously generated oxygen radicals, replication forks encountering single stranded DNA breaks or other DNA lesions, or exogenous agents such as ionizing radiation or chemotherapeutic drugs [3]. There are two main options for repairing a DSB: homologous recombination (HR) and non-homologous end joining (NHEJ) [3]. The focus of this paper is proteins, specifically RecA and SSB, involved in homologous recombination in E. coli.
RecA

RecA is a 38kDA ATPase, a class of enzymes that catalyze the hydrolysis of ATP, found in Escherichia coli that is essential for DNA repair and maintenance [2].
In order to function, the must bind at the site of DNA damage. This occurs in two distinct stages:
- Nucleation or the slow aggregation and binding of a few RecA molecules to the DNA lesion.
- Growth or the rapid binding of successive RecA proteins in competition with single stranded DNA-binding (SSB) protein to coat the entire lesion.
ATP and ssDNA bind at the RecA-RecA interface cooperatively; together, the complex forms a helical filament which binds dsDNA, searches for homology, and then catalyses strand exchange [2].
Single Strand DNA Binding Protein
In E. coli, the single-stranded DNA-binding protein (SSB) is homotetramer consisting of 18,843 kDa subunits which binds tightly and cooperatively to ssDNA. It is involved in most aspects of DNA manipulation: replication, repair, and recombination. Its functions include prevention of reannealing of ssDNA, protection of ssDNA from nuclease digestion, enhancement of helix destabilization by DNA helicases, primosome assembly, enhancement of replication fidelity [4].
In DNA damage, SSB is involved in induction of the SOS response and recombination repair. During recombination, SSB promotes he binding of RecA protein and strand uptake [4].
RecO, RecR, and RecF

RecF, RecO, and RecF are three of a dozen proteins involved in the RecF pathway of recombination usually reserved for ssDNA gap repair. Although other proteins in the pathway have well defined and well understood roles in recombination process the roles of RecF, RecO, and RecR proteins remain unclear [8].
Studies show that RecF, recO, and RecR act in concert to facilitate the loading of RecA protein onto SSB-coated gDNA. First, RecF binds to the dsDNA-ssDNA junction through recognition of the 5’ end of the dsDNA [8]. Then RecOR binds to the DNA-RecF complex and the assembly of the RecFOR complex on the DNA causes conformational changes of the ssDNA-SSB complexes and/or displaces some SSB protein allowing for nucleation of RecA to occur [8].
Introduction
The purpose of this study is to develop methods to visualize and to define the different stages of the binding of RecA to a SSB coated ssDNA molecule. By adjusting the conditions in which the RecA-ssDNA filament formation occurs, the researchers hope to better understand the mechanisms and rates of this process and roles of various compounds and mediators play in RecA-ssDNA filament formation.
Methods


In order to visualized ssDNA, researchers tagged the 3’ ends of bacteriophage lambda DNA (48.5 kb) with biotin. Then, they denatured it into single strands before coating the strands with fluorescence tagged single strand binding protein (SSB). These coated ssDNA was then attached to a glass cover slip covered with streptavidin, a protein which binds strongly to the biotin tag, of a flow chamber. They where then able to visualize the coated strand using total internal reflection fluorescence (TIRF) microscopy (Figure 1a/b, top panel). The tagged SSB was then replaced with wildtype SSB (Figure 1a/b, second panel). The researchers were then able to flow solutions containing fluorescent RecA protein, fluorescein-RecA, through the chamber, over the ssDNA and visualize the binding of RecA to the tethered ssDNA (Figure 1a/b, remaining panels).
In order to test the effect of RecF, RecO, and RecR, the researchers needed to generate dsDNA with a ssDNA gap because it has been shown that RecF functions in the presence of only a dsDNA-ssDNA junction [8]. They generated a linear strand of dsDNA with an 8 kilobases ssDNA gap which had biotin tags on either end. They bound either end of a single gapped-DNA molecule to streptavidin coated beads (Figure 4g, top panel). A split beam optical trap was used to trap the beads and rotate the DNA perpendicular to the flow of an adjoined flow cell and to dip the strand into the reaction chamber. The binding of the molecules was measured using the fluorescent microscope (Figure 4g, bottom panel) which was coupled to the split beam optical trap.
Results
In this study, a method to directly image the formation of the RecA-ssDNA filament was developed (see methods for details). Using this visualization technique, the nucleation rate of RecA on a SSB-coated ssDNA molecule was measured. The researchers plotted the clusters of RecA versus time to give the nucleation rate at different concentrations of RecA ranging from 100nM to 450 nM and concluded that the nucleation rate increased as the concentration of RecA was increased (Left graph below).
Since individual clusters of RecA were visible, researchers were able to plot the growth of each cluster in order to establish a growth rate at various concentrations of RecA by plotting length of RecA-ssDNA filament by time. They were then able to extrapolate the lag time, or the time it takes for the nucleation to occur (right graph above).
After establishing growth rate, they determined directionality of growth. First by completing the nucleation step with Cy3 tagged RecA (Figure 3b, second panel, red) and then using fluorescein-RecA (green) to track growth of those clusters. Observations indicate that 5’→3’ direction was preferred, but that bidirectional growth was possible.
They then tested various conditions to see their effect on the nucleation and growth rates. First, they modulated the concentration of magnesium acetate ([Me(Oac)2), which associates with ATP. As the concentration of Me(Oac)2 was increased the rate of nucleation increased, but it had no effect on growth rate suggesting that the presence of Mg+-ATP cofactor is critical for increasing the rate of nucleation of RecA, but not in growth rate. The effects of ATP hydrolysis was tested using ATP, ATP-γ-S (an non-hydrolyzable analog), and dATP (a more readily hydrolysable version of ATP). In the presence of ATP-γ-S and dATP, the nucleation rate is 10-fold and 5-fold faster, respectively, than with ATP, suggesting that hydrolysis has no bearing on the stability of the filament. The growth rate was not statistically effected by ATP-γ-S or dATP compared to ATP. These results suggest that ATP acts as a conformational regulator, increasing RecA protein’s affinity for ssDNA upon binding.
Next, the effect of mediator proteins were tested, specifically RecO, RecR, and RecF, genetically and biochemically established mediator proteins in E. coli [1]. They tested two combinations: the RecOR complex which interacts with SSB to mediate assembly in a structure-independent manner, and RecFOR complex which acts at the junction of ssDNA and dsDNA [1]. To test the effect of these cofactors, a new set up was used described in the Methods section above. In the presence of RecOR, the mean lag time decreased, some occurring in less time than RecA alone. The addition of RecF further decreased the mean lag time (Figure 4h, below). RecOR accelerated the growth rate but RecF had no effect (Figure 4i, below).
Based on these results, the researches created a model (Figure 4j, below) in which RecOR interacts with SSB to facilitate RecA assembly [1,7] by trapping transiently unbound ssDNA thus weakening the SSB-ssDNA binding and facilitating RecA binding [1].
Significance/Applications
The use of this approach to establish that mediators such as RecOR and RecF can effect the rate limiting step in biological processes such nucleation, enhance filament growth, or both suggests it may be applicable in other, similar systems. For exampled, it could be used to study the nucleation and growth of Rad51, the human othologue of RecA, and DNA filaments; the filament formation is regulated by BRCA2 which could be studies in a similar manner to the roles of RecOR and RecF. The technical approach could also be used to understand the role of RAD51 paralogues in recombination-mediated DNA repair [1].
References
- Bell, Jason C., Plank, Jody L., Dombrowski, Christopher C., and Kowalczykowski. Direct imaging of RecA nucleation and growth on single molecules of SSB-coated ssDNA. Nature 491, 274 - 278 (2012).
- Chen, Z., Yang, H., Pavletich, N.P. Machanism of homolgous recombination from the RecA-ssDNA/dsDNA strucures. Nature 435. 489-494 (2008)
- Khanna, K. K. & Jackson, S. P. DNA double-strand breaks: signaling, repair and the cancer connection. Nature Genetics 27, 247 - 254 (2001)
- Meyer, R. R. & Laine, P. S. The single-stranded DNA-binding protein of Escherichia coli. Microbiology Review 54, 342-380 (1990)
- Cox, Michael M. Motoring along with the bacterial RecA. Nature Reveiws Molecular Cell Biology 8, 127-138 (2007)
- Kowalczykowski, S. C., Clow, J., Somani, R. & Varghese, A. Effects of Escherichia coli SSB protein on the binding of Escheria coli RecA protein to single-stranded DNA. Demonstraion of competitive binding and the lack of a specific protein-protein interaction. J. Mol. Biol. 193, 81-95 (1987)
- Umezu, K., Chi, N. W. & Kolodner, R. D. Biochemical interaction of the Escherichia coli RecF, RecO, and RecR proteins with RecA protein and single-stranded DNA binding protein. Proc. Natl Acad. Sci. USA 90, 3875–3879 (1993).
- Morimatsu, Katsumi & Kowalczykowski, Stephen C. RecFOR Proteins Load RecA Protein onto Gapped DNA to Accelerate DNA Strand Exchange: A Universal Step of Recombinational Repair. Molecular Cell 11, 1337-1347 (2003).