CH391L/S13/In vitro Selection of FNAs
Introduction
Functional nucleic acids (FNAs, as described by Dr. S.K. Silverman, are DNA and RNA aptamers that bind targets, or they are deoxyribozymes (single stranded DNA) and ribozymes (RNA) that have catalytic activity. Aptamers, Ribozymes, and Deoxyribozymes are grouped into three main categories that are further classified into either natural or artificial depending on their origin. The exception being Deoxyribozymes as they have yet to be discovered in a living organism. Although the first ribozyme was discovered only in the 1980s, the search for new and better FNAs continues. This has led the discovery of new methods, such as the SELEX [1, 2] or In vitro selection process, as we strive to their potential both as tools for exploring biology and solving real world problem solving.
In vitro Selection of Functional Nucleic Acids

The figure to the right, SELEX or In vivo Selection Experiment , describes the basic method for performing a SELEX or In vivo selection experiment using single stranded nucleic acids that are chemically synthesized,with a constant region (CR) and a random region. The first step is subjecting the population of single stranded nucleic acids to specific selective condition in which function is possible. Then a (2) diverse subset of the population will perform the desired function and will be then (3) PCR (Polymerase Chain Reaction) amplified to generate double stranded nucleic acids making use of CR introduced previously. The previous step is necessary for the selection's continuation into the next selection round, while at the same time a sample is obtained and can be sequenced.
Functional Nucleic Acids

Ribozymes
As previously mentioned ribozymes fall under the category of enzymes. Most of the ribozymes studied up until recently in living organism fall into 9 classes. Of these most perform some type scission and ligation reaction. In the case of in vitro selected ribozymes their function has been expanded to include . It was in such attempts that TPP ribozyme biobrick was made from both by combining an aptamer and a ribozyme for regulatory purposes. Most of other focus also on the regulation of an specific part, the self-modification of the part itself or both. Finally, one of the newest tools available to new tool are flexizymes that perform a self-aminoacylating reaction on an in vitro selected tRNA with a N70 region and that can add nonnatural amino acids by reprogramming genetic code[8].
Natural Ribozymes
- Splicing Ribozymes-
- Riboswitches - Translational control mechanism found at the mRNA level.
Deoxyribozymes
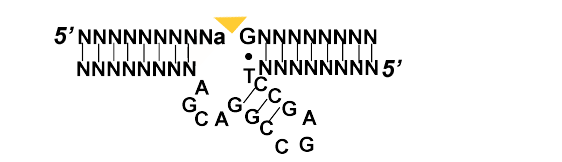
An interesting discovery was made in the early 1990s, when for the first time DNA was shown that, besides being a genetic information storage molecule, it could also be both an enzyme and an aptamer. In the figure below you can observe 8-17, an RNA cleaving DNA enzyme. This molecule with 10-23 were the first to be described and tested in vivo as potential new therapies for cleaving the expressed mRNA of a virus. [6] Although proteins offer a larger diversity chemistries, depending on amino acids vs. both kinds of nucleic acids, as the latter ones depend on a limited array of nucleotides. Until know around a dozen distinct types of reactions. These include the following activities such as self-phosphorylation, RNA labeling, depurination,etc [9]
Aptamers and Riboswitches
The word aptamer from the latin aptus and translates as the past participle of to fit were originally identified by employing the protocol SELEX. Therefore the word Aptamer describes their basic function as RNA or single stranded DNA (ssDNA)that can bind a ligand by assuming an specific structure.[10, 11] Yet, it would take several years until the discovery of the first in vivo aptamer or riboswitch [12]. See the following page to get a better understanding of aptamers and riboswitches.
Conclusion
References
<biblio>
- Cech1982 pmid=6297745
- Altman1983 pmid=6197186
- Breaker2002 pmid=12022469
- Silverman2009 isbn=978-0-387-73711-9
- Wilson1999 pmid=10872462
- Ellington1990 pmid=1697402
- Gold1990 pmid=2200121
- Winkler2002 pmid=12410317
- Zucker2003 pmid=12824337
- GoTo2006 pmid=17150804
- Patel2007 pmid=17846637
- Breaker2000 pmid=11187837
<\biblio>