CH391L/S13/In vitro Selection of FNAs
Introduction
Although the vast majority of such ligand binding activities or enzymatic activities known are performed by proteins a secondary subset of these fall under the category of Functional nucleic acids (FNAs. FNAs are RNA, ssDNA, or XNA(nucleic acid analogues) that perform an activity such as binding or catalyzing a reaction. FNAs are grouped into three main categories Aptamers, Ribozymes, and Deoxyribozymes that are further classified into either natural or artificial depending on their origin. The exception being Deoxyribozymes as they have yet to be discovered in a living organism. Still, It was only in the 1980s that the 1st ribozyme was discovered that we started to study FNAs and have allowed for the discovery of new methods, such as the SELEX or In vitro selection process that we are expanding their potential both as tools for exploring biology and real world problem solving.
<!Note to self: mention RNA World>
Functional Nucleic Acids
Ribozymes
As previously mentioned ribozymes fall under the category of enzymes. Most of the ribozymes studied up until recently in living organism fall into 9 classes.
- Cleaving Ribozymes
- 1 Self-cleaving i.e. The Hammerhead family (satellite RNA)
- 2 Trans-cleaving i.e. RNase P
- Splicing Ribozymes
- 3 Group I
- 4 Group II
- Riboswitches
- 5 Thermosensors
- 6 sRNAs
- 7 T-Boxes
- 8 Metabolites
- 9 Magnesium
Deoxyribozymes
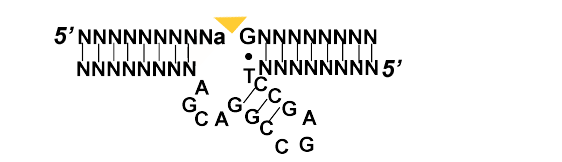
Aptamers and Riboswitches
The word aptamer from the latin aptus and translates as the past participle of to fit were originally identified by employing the protocol SELEX. Therefore the word Aptamer describes their basic function as RNA or single stranded DNA (ssDNA)that can bind a ligand by assuming an specific structure.[5, 6] Yet, it would take several years until the discovery of the first in vivo aptamer or riboswitch [7]. See the following page to get a better understanding of aptamers and riboswitches.
In vitro Selection of Functional Nucleic Acids

The image presented describes the basic method for performing a SELEX or In vivo selection experiment using single stranded nucleic acids (RNA,ssDNA,XNA) that are chemically synthesizedan have a constant region (CR) and a random region. Having the CR allows later amplification using PCR. The first step is subjecting the population of single stranded nucleic acids to specific selective condition in which function is possible. Then a (2) diverse subset of the population will perform the desired function and will be then (3) PCR amplified to make double stranded nucleic acids with the use of the CR introduced previously. Therefore the selection can continue to a following round, while at the same time a sample is obtained and can be sequenced. [8]
New Methods and Tools that assit In vitro selections
Computational Methods
- Mfold - originally developed as a GUI or graphical user interface server for the prediction of the secondary structure of single stranded nucleic acids. The core algorithm predicts structures based on minimum free energies necessary to fold a structure. This algorithm can be used initially as a step to predict possible secondary structures of active sequences [9].
FRET
- Fluorescent analogues
Next Generation Sequencing
New Applications of FNAs
<biblio>
- Cech1982 pmid=6297745
- Altman1983 pmid=6197186
- Breaker2002 pmid=12022469
- Silverman2009 isbn=978-0-387-73711-9
- Wilson1999 pmid=10872462
- Ellington1990 pmid=1697402
- Gold1990 pmid=2200121
- Winkler2002 pmid=12410317
- Zucker2003 pmid=12824337
<\biblio>