CH391L/S13/GeneticMarkers: Difference between revisions
No edit summary |
No edit summary |
||
| Line 12: | Line 12: | ||
[[Image:amp.png|thumb|center|500px| The structure of ampicillin, including the central beta-lactam ring cleaved by the ''ampR'' gene product [http://en.wikipedia.org/wiki/Ampicillin Ampicillin on Wikipedia]]] | [[Image:amp.png|thumb|center|500px| The structure of ampicillin, including the central beta-lactam ring cleaved by the ''ampR'' gene product [http://en.wikipedia.org/wiki/Ampicillin Ampicillin on Wikipedia]]] | ||
The common ''ampR'' gene, as used in the ''E. coli'' pBR322 plasmid, was naturally derived from ''Salmonella'' bacteria through transposition. Because it efficiently cleaves amp and carb, it's one of the most useful markers in ''E. coli''. One downside to the use of ''ampR'' is that removal of antibiotic from selective media by beta-lactamase can allow for growth of cells that lack resistance. This is often witnessed in the form of satellite colonies on amp plates.<cite>Sutcliffe78</cite> | The common ''ampR'' gene, as used in the ''E. coli'' pBR322 plasmid, was naturally derived from ''Salmonella'' bacteria through transposition. Because it efficiently cleaves amp and carb, it's one of the most useful markers in ''E. coli''. One downside to the use of ''ampR'' is that removal of antibiotic from selective media by beta-lactamase can allow for growth of cells that lack resistance. This is often witnessed in the form of satellite colonies on amp plates.<cite>Sutcliffe78</cite> <br> | ||
For | For information on other common antibiotic selection markers, see the links below: <br> | ||
[http://en.wikipedia.org/wiki/Kanamycin Kanamycin on Wikipedia] <br> | [http://en.wikipedia.org/wiki/Kanamycin Kanamycin on Wikipedia] <br> | ||
[http://en.wikipedia.org/wiki/Chloramphenicol Chloramphenicol on Wikipedia] <br> | [http://en.wikipedia.org/wiki/Chloramphenicol Chloramphenicol on Wikipedia] <br> | ||
Revision as of 11:29, 4 March 2013
Introduction
The ability to introduce exogenous DNA into an organism to alter its genetic program is one of the most crucial tools in modern biology. Early work showed that certain bacteria could acquire the traits of a related strain through the addition of heat-killed cells. Although it was not well understood at the time, the transfer of gene-encoding DNA from one strain to another facilitated this. This concept was turned into a useful tool upon the advent of bacterial plasmid transformations in the early 1970's, which allowed genes of interest to be easily inserted into E. coli. Over the years, methods have been developed to introduce exogenous genes into a wide range of useful organisms, including bacteria, yeasts, plants, and animal tissues. These methods vary enormously in efficiency however, necessitating a way to identify and isolate cells which contain the DNA of interest. This can be accomplished either by screening for successfully modified cells, or through selection.
Screening vs. Selection
In the case of bacterial transformations, it's possible to plate cells post-transformation on non-selective media and screen each individual colony to identify those that have been successfully modified. Because each colony arises from a single cell present during transformation, the colony is made up of a set of identical clones. Screening can involve testing for the desired DNA itself or for the product of an inserted gene. This process is extremely difficult however, as the vast majority of cells will not be successfully transformed, so many colonies need to be tested to identify even a single transformant. A much more efficient strategy is to use a selectable genetic marker that allows only those cells which have been transformed to survive under certain growth conditions. These marker genes may be combined with the DNA of interest on a single plasmid, thus ensuring that any cells that survive selection contain the gene(s) of interest. The most commonly used selectable markers in bacteria are genes that provide resistance to a specific antibiotic upon transformation, allowing for positive selection of cells containing the marker.
Antibiotic Resistance Markers: ampR
Perhaps the most commonly used selectable marker in bacteria is the ampR gene, which provides resistance to certain beta-lactam antibiotics such as ampicilllin (amp) and its more stable relative carbenicillin (carb). Beta-lactam antibiotics are penicillin derivatives that inhibit synthesis of bacterial peptidoglycan cell walls, arresting cell division and ultimately leading to cell death. Although beta-lactams are primarily functional against gram-positive bacteria due to their larger cell walls, some examples, such as amp and carb, are capable of killing gram-negative E. coli. All antibiotics in the family share a central four atom ring structure known as the beta-lactam ring, which serves as a cleavage target for enzymes known as beta-lactamases. Due to the shared structural features of becta-lactam antibiotics, these enzymes often have promiscuous activities that target multiple drugs. Upon cleavage of the lactam ring, antibiotics such as amp lose their toxicity, allowing cell growth and division to resume.[1]
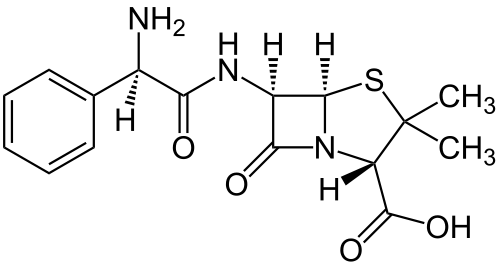
The common ampR gene, as used in the E. coli pBR322 plasmid, was naturally derived from Salmonella bacteria through transposition. Because it efficiently cleaves amp and carb, it's one of the most useful markers in E. coli. One downside to the use of ampR is that removal of antibiotic from selective media by beta-lactamase can allow for growth of cells that lack resistance. This is often witnessed in the form of satellite colonies on amp plates.[1]
For information on other common antibiotic selection markers, see the links below:
Kanamycin on Wikipedia
Chloramphenicol on Wikipedia
Antibiotic/Non-Antibiotic Dual Selection: tetA(C)
Tetracycline Positive Selection
The tetA(C) gene is primarily used for positive selection in bacteria, similar to ampR. tetA(C) encodes a membrane-bound transporter that rapidly pumps the antibiotic tetracycline out of bacterial cells. This process is energy dependent, as the protein uses the influx of H+ ions from the surrounding environment to drive the process. Tetracycline is a broad-spectrum, polyketide antibiotic derived from Streptomyces that inhibits bacterial translation. The antibiotic binds the 30s subunit of bacterial ribosomes, blocking entry of aminoacyl-tRNAs to the A site of the ribosome.[2]

Because tetracycline resistance is gained through expression of a transporter and not a modifying enzyme, selective media maintains its antibiotic levels during growth of resistant cells. High levels of tetA expression can have a detrimental effect on the cells however, due the energy cost of pumping tetracycline and increased vulnerability to certain extracellular conditions. [3]
Nickel Salt Negative Selection
Expression of tetA in bacteria has the side effect of making cells more vulnerable to poisoning by either lipophilic chelating agents or metal salts. Although tetA expressing Salmonella strains were shown to be extremely sensitive to chelating agents such as fusaric or quinaldic acids, these compounds are only marginally effective on E. coli. Based on earlier observations that metals like cadmium inhibited growth of tetracycline-resistant bacteria, a technique was developed that uses nickel salts to select against tetA expressing E. coli with much greater efficiency. Cheap, non-toxic Nickel Chloride is the most commonly used selection agent. Although originally used to select for cells that had lost a tetA marker, this method has proven useful in dual selection schemes for evolving regulators of gene expression. [3, 4]
Engineering Riboswitches using tetA Dual Selection
The evolution of functional Riboswitches requires selection of a library of mutants in both an ON state and an OFF state. This removes variants that either activate or repress gene expression regardless of the small molecule used to regulate them. Previous attempts at dual selection required the use of both positive and negative selection markers. This necessitated intermediate steps to purify plasmids from the pool and re-transform them along with either the positive or negative marker. Aside from being labor intensive, this method also increased the rate of false positives in the pool [5, 6].
The fact that a single tetA gene can be used for both positive and negative selection greatly simplifies the process of selecting for riboswitches, as a culture of cells expressing a library of riboswitches can be alternately swapped between media that selects for or against expression of the tetracycline transporter. This strategy was used by the Yokobayashi Lab at UC Davis to select for mutants of the E. coli TPP riboswitch (from thiM) that activated downstream gene expression in the presence of thiamine instead of repressing it. A library of TPP riboswitch variants was cloned upstream of a tetA marker and transformed into E. coli. The first round of selection involved plating transformants on media with thiamine and tetracycline to select for mutants that were turned ON. The survivors were then transferred to media with nickel chloride and no thiamine to remove any variants that did not turn off in the absence of thiamine. A similar approach can be carried out in liquid culture, as shown below.[4].

Using a tetA Fusion Protein for Monitoring Selection
More recent work by the Yokobayashi Lab involved combining the tetA marker with GFP to allow screening of thiM riboswitch function following each round of dual selection. The tetA transporter remains functional when GFP is fused to its c-terminal end, facilitating selection and screening steps with the same gene product. To measure function of different riboswitch variants in the selection pool, samples were plated on LB and individual colonies were picked and used to seed culture in non-selective media. GFP fluorescence of each culture was measured to determine the amount of gene expression generated by that particular riboswitch, and functional variants were sequenced[6].
Non-Antibiotic Selection: GFAT
Glutamine:fructose-6-phosphate aminotransferase (GFAT) is an enzyme required for the biosynthesis of hexosamines in both bacteria and eukaryotes. GFAT catalyzes the rate-limiting first step in the pathway, converting fructose-6-phosphate into glucosamine-6-phosphate. In E. coli, GFAT is expressed from the glmS gene, which is necessary for growth in minimal media. Recent work has shown that E. coli lacking glmS are capable of growing at near wild type levels when supplemented with glucosamine however, and complementation with a plasmid-encoded glmS gene restores hexosamine synthesis and growth[7].
Selection of glmS expressing cells is complicated by the fact that rich media such as LB contains sufficient glucosamine to support limited growth of glmS knockout cells. Growth tests using M9 minimal media with individual LB components added to it showed that yeast extract is the source of glucosamine that allows growth of the knockout strain. Thus, selections with GFAT markers are limited to minimal media, but the addition of tryptone increases growth rates while still allowing selection. Under these conditions, selection for glmS was shown to be an effective replacement for ampR selection[7].
Future Directions
Although the currently available set of selectable markers has helped us to overcome many of the challenges involved in synthetic biology, the continued discovery of new markers may greatly simplify the process of designing organisms with useful functions. The variety of useful markers available for engineering of higher eukaryotes, including animals and human tissue culture lines pales in comparison to those available for use in bacteria or even yeast. Thus, the discovery of new selection schemes may allow experiments that are not currently feasible.
Another area of ongoing progress is the discovery of non-antibiotic markers to potentially replace commonly used genes like ampR in bacterial engineering. Although antibiotic resistance is an increasingly common trait in wild bacteria, the possibility of engineered organisms transferring their resistance to otherwise susceptible bacteria is a frequent cause for alarm. If engineered bacteria are to prove useful for tasks like bio-remediation, we will have to engineer them without the benefit of antibiotic selection or markers that require growth in minimal media.
See Also:
Selectable Genetic Markers
Toxin/Antitoxin Systems
Counterselection
iGEM Connections
The 2011 Peking_R iGEM team from Peking University used tetA dual selection to engineer a ribozyme-based "genetic rheostat." Their approach involved fusing the TPP aptamer domain from the thiM riboswitch to a section of the hammerhead ribozyme. Portions of the construct were randomized and selections were carried out in a similar fashion to those performed by the Yokobayashi Lab[6]. This approach did increase the ratio of ON/OFF ribozymes in their pool, and similar selections using an adenine aptamer domain also demonstrated the potential of the approach.
References
- Sutcliffe JG. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3737-41. DOI:10.1073/pnas.75.8.3737 |
Background on the ampR gene from pBR322
- McNicholas P, Chopra I, and Rothstein DM. Genetic analysis of the tetA(C) gene on plasmid pBR322. J Bacteriol. 1992 Dec;174(24):7926-33. DOI:10.1128/jb.174.24.7926-7933.1992 |
The tetA(C) gene from pBR322
- Podolsky T, Fong ST, and Lee BT. Direct selection of tetracycline-sensitive Escherichia coli cells using nickel salts. Plasmid. 1996 Sep;36(2):112-5. DOI:10.1006/plas.1996.0038 |
Nickel selection with tetA
- Nomura Y and Yokobayashi Y. Reengineering a natural riboswitch by dual genetic selection. J Am Chem Soc. 2007 Nov 14;129(45):13814-5. DOI:10.1021/ja076298b |
Reengineering the TPP riboswitch using tetA dual selection
- Collins CH, Leadbetter JR, and Arnold FH. Dual selection enhances the signaling specificity of a variant of the quorum-sensing transcriptional activator LuxR. Nat Biotechnol. 2006 Jun;24(6):708-12. DOI:10.1038/nbt1209 |
Dual selection with separate markers
- Muranaka N, Sharma V, Nomura Y, and Yokobayashi Y. An efficient platform for genetic selection and screening of gene switches in Escherichia coli. Nucleic Acids Res. 2009 Apr;37(5):e39. DOI:10.1093/nar/gkp039 |
Riboswitch selection/screening using a tetA-GFP fusion marker
- Wu G, Sun Y, Qu W, Huang Y, Lu L, Li L, and Shao W. Application of GFAT as a novel selection marker to mediate gene expression. PLoS One. 2011 Feb 14;6(2):e17082. DOI:10.1371/journal.pone.0017082 |
GFAT as a selectable marker