CH391L/S13/DnaAssembly
DNA assembly is a key component to synthetic biology.
A large number of parts have been made by the synthetic biology community. Many can be found as part of the Registry of Standard Biological Parts. These modular genetic components are designed to be easy to acquire and assemble to facilitate the building of more complex biological devices. To learn more about the Registry and the biological parts known as BioBricks™, see the entry for the iGEM Registry.
The Registry of Standard Biological Parts is an attempt to create an annotated and characterized repository of biological parts. It is motivated in part because synthetic biologists rely on the ability to make testable biological units. This means that the ability to gather, manipulate, or even create genetic material is vital for "doing" synthetic biology. This page details how to create DNA from small (<60 nts) oligonucleotides to larger genes (~400 nts) to genome sized (~500 ,000 nts) biological units.
DNA Synthesis
Oligonucleotide Synthesis
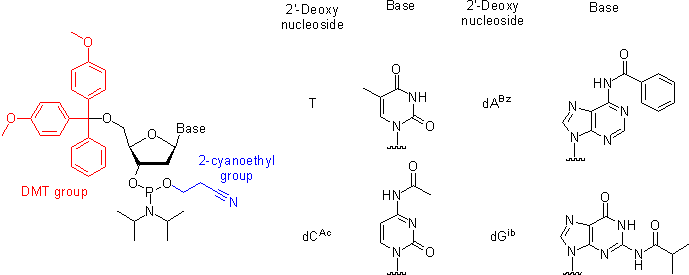
Oligonucleotides are chemically synthesized from DNA phosphoramidite monomers. Briefly, activated phosphoramidite monomers are added in the 3' to 5' direction using a cyclical activation and blocking chemistry to obtain a DNA polymer linked by phosphodiester bonds.
Chemical synthesis is currently limited to oligonucleotides of about 200 nt in length.
Gene Synthesis
Gene synthesis, or artificial gene synthesis, refers to the process of creating a nucleic acid template for a gene in vitro, without the requirement of a preexisting DNA template. Soon after the elucidation of the genetic code and the description of the central dogma of molecular biology, there arose a need to synthesize genes de novo in order to study their biological function both in the test tube and in model organisms. Chemical synthesis of DNA has grown from an expensive and time-consuming process into a viable commercial industry capable of high-throughput manufacture of almost any scale of custom DNA molecules in almost any context. This allows species-specific gene optimization, creation of genes from rare or dangerous sources, and combinatorial assembly of any DNA sequence that can be chemically synthesized, even including non-traditional bases. The most advanced applications of gene synthesis have been applied to the recent creation of completely synthetic minimal genomes in prokaryotes.
Despite nearly four decades of progress in gene synthesis technologies, most DNA sequences used in modern molecular biology are assembled in part or in whole from naturally occurring templates. However this limits the scope and applications to previously existing genes and the results of large-scale genomic surveys of novel genes from nature. Modern gene synthesis relies heavily on advancements in chemical DNA oligonucleotide synthesis, with the primary challenges being scale, cost, fidelity and the eventual assembly of complete gene products.
A directory of commercial gene synthesis providers can be found at Genespace. The company biomatik is not included on this list.
History of Gene Synthesis
Gene synthesis predates the invention of restriction enzymes and molecular cloning techniques by several years. The first gene to be completely synthesized in vitro was a 77-nt alanine transfer RNA by the laboratory of Har Gobind Khorana in 1972 [1]. This was the result of nearly five years of work and resulted in a DNA template without promoter or transcriptional control sequences. The first peptide- and protein-producing synthetic genes were created in 1977 and 1979, respectively [2, 3]. Steady advancement has led to recent synthesis of complete gene clusters tens of thousands of nucleotides in length, and ultimately a bacterial genome approximately 1.2 million bases in length.
Cost Analysis



References
Cost
Time
Molecular Cloning
Restriction Enzyme
BioBricks
BglBricks
list of BioBrick Foundation Standards
Polymerase Chain Reaction
TOPO TA cloning (invitrogen)
SOE (splice by overlap extension) pcr
PCA
Ligation
Recombination/Homology
j5 from Nathan Hillson
-In-Fusion (Clontech) poxvirus DNA polymerase with 3′–5′ exonuclease activity
-In-Fusion BioBrick Assembly
-cold fusion (SBI)
-golden gate
-MoClo [4]
-GoldenBraid
-SLIC sequence and ligation independent cloning T4 DNA polymerase (exonuclease)
-Gibson T5 exonuclease, Phusion polymerase, Taq ligase
-CPEC circular polymerase extension cloning
-SLiCE (Seamless Ligation Cloning Extract) in vitro homologous recombination
Uncategorized
-Fast Seamless Cloning (Dogene)
-CloneEZ kit (Genescript)
-GENEART
-lic
-gateway
-lambda red
Transformation
- e coli
- s. cerevisiae
Bacterial Mating
MAGIC
More cloning strategies found here
References
- Weber E, Engler C, Gruetzner R, Werner S, and Marillonnet S. A modular cloning system for standardized assembly of multigene constructs. PLoS One. 2011 Feb 18;6(2):e16765. DOI:10.1371/journal.pone.0016765 |
MoClo
- Hughes RA, Miklos AE, and Ellington AD. Gene synthesis: methods and applications. Methods Enzymol. 2011;498:277-309. DOI:10.1016/B978-0-12-385120-8.00012-7 |
Gene Synthesis Review
- Werner S, Engler C, Weber E, Gruetzner R, and Marillonnet S. Fast track assembly of multigene constructs using Golden Gate cloning and the MoClo system. Bioeng Bugs. 2012 Jan 1;3(1):38-43. DOI:10.4161/bbug.3.1.18223 |
MoClo
- Sarrion-Perdigones A, Falconi EE, Zandalinas SI, Juárez P, Fernández-del-Carmen A, Granell A, and Orzaez D. GoldenBraid: an iterative cloning system for standardized assembly of reusable genetic modules. PLoS One. 2011;6(7):e21622. DOI:10.1371/journal.pone.0021622 |
GoldenBraid
- Engler C, Kandzia R, and Marillonnet S. A one pot, one step, precision cloning method with high throughput capability. PLoS One. 2008;3(11):e3647. DOI:10.1371/journal.pone.0003647 |
GoldenGate
- Quan J and Tian J. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS One. 2009 Jul 30;4(7):e6441. DOI:10.1371/journal.pone.0006441 |
CPEC
- Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley A, Thomas DW, Algire MA, Merryman C, Young L, Noskov VN, Glass JI, Venter JC, Hutchison CA 3rd, and Smith HO. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008 Feb 29;319(5867):1215-20. DOI:10.1126/science.1151721 |
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, and Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009 May;6(5):343-5. DOI:10.1038/nmeth.1318 |
T5 exonuclease recombination
- Li MZ and Elledge SJ. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods. 2007 Mar;4(3):251-6. DOI:10.1038/nmeth1010 |
SLIC
- Li MZ and Elledge SJ. SLIC: a method for sequence- and ligation-independent cloning. Methods Mol Biol. 2012;852:51-9. DOI:10.1007/978-1-61779-564-0_5 |
SLIC
- Sleight SC, Bartley BA, Lieviant JA, and Sauro HM. In-Fusion BioBrick assembly and re-engineering. Nucleic Acids Res. 2010 May;38(8):2624-36. DOI:10.1093/nar/gkq179 |
In-Fusion biobrick
- Zhu B, Cai G, Hall EO, and Freeman GJ. In-fusion assembly: seamless engineering of multidomain fusion proteins, modular vectors, and mutations. Biotechniques. 2007 Sep;43(3):354-9. DOI:10.2144/000112536 |
in-fusion
- Benoit RM, Wilhelm RN, Scherer-Becker D, and Ostermeier C. An improved method for fast, robust, and seamless integration of DNA fragments into multiple plasmids. Protein Expr Purif. 2006 Jan;45(1):66-71. DOI:10.1016/j.pep.2005.09.022 |
in-fusion
- Geu-Flores F, Nour-Eldin HH, Nielsen MT, and Halkier BA. USER fusion: a rapid and efficient method for simultaneous fusion and cloning of multiple PCR products. Nucleic Acids Res. 2007;35(7):e55. DOI:10.1093/nar/gkm106 |
USER
- Gibson DG. Synthesis of DNA fragments in yeast by one-step assembly of overlapping oligonucleotides. Nucleic Acids Res. 2009 Nov;37(20):6984-90. DOI:10.1093/nar/gkp687 |
oligonucleotide assembly
- Horton RM, Cai ZL, Ho SN, and Pease LR. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 1990 May;8(5):528-35.
SOEing
- Czar MJ, Anderson JC, Bader JS, and Peccoud J. Gene synthesis demystified. Trends Biotechnol. 2009 Feb;27(2):63-72. DOI:10.1016/j.tibtech.2008.10.007 |
review
- Stemmer WP, Crameri A, Ha KD, Brennan TM, and Heyneker HL. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene. 1995 Oct 16;164(1):49-53. DOI:10.1016/0378-1119(95)00511-4 |
PCA
- Aslanidis C and de Jong PJ. Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res. 1990 Oct 25;18(20):6069-74. DOI:10.1093/nar/18.20.6069 |
LIC
- Aslanidis C, de Jong PJ, and Schmitz G. Minimal length requirement of the single-stranded tails for ligation-independent cloning (LIC) of PCR products. PCR Methods Appl. 1994 Dec;4(3):172-7. DOI:10.1101/gr.4.3.172 |
LIC
- Li C and Evans RM. Ligation independent cloning irrespective of restriction site compatibility. Nucleic Acids Res. 1997 Oct 15;25(20):4165-6. DOI:10.1093/nar/25.20.4165 |
LIC
- Angrand PO, Daigle N, van der Hoeven F, Schöler HR, and Stewart AF. Simplified generation of targeting constructs using ET recombination. Nucleic Acids Res. 1999 Sep 1;27(17):e16. DOI:10.1093/nar/27.17.e16 |
lambda Red recombinase
- Hartley JL, Temple GF, and Brasch MA. DNA cloning using in vitro site-specific recombination. Genome Res. 2000 Nov;10(11):1788-95. DOI:10.1101/gr.143000 |
Gateway lambda Int
- Khalil AM, Julius JA, and Bachant J. One step construction of PCR mutagenized libraries for genetic analysis by recombination cloning. Nucleic Acids Res. 2007;35(16):e104. DOI:10.1093/nar/gkm583 |
Gateway lambda Cre
- Larionov V, Kouprina N, Graves J, Chen XN, Korenberg JR, and Resnick MA. Specific cloning of human DNA as yeast artificial chromosomes by transformation-associated recombination. Proc Natl Acad Sci U S A. 1996 Jan 9;93(1):491-6. DOI:10.1073/pnas.93.1.491 |
Transformation-associated recombination (TAR) cloning
-
j5 DNA Assembly Design Automation Software doi: 10.1021/sb2000116
- Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang RY, Algire MA, Benders GA, Montague MG, Ma L, Moodie MM, Merryman C, Vashee S, Krishnakumar R, Assad-Garcia N, Andrews-Pfannkoch C, Denisova EA, Young L, Qi ZQ, Segall-Shapiro TH, Calvey CH, Parmar PP, Hutchison CA 3rd, Smith HO, and Venter JC. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010 Jul 2;329(5987):52-6. DOI:10.1126/science.1190719 |
genome replacement
- Li MZ and Elledge SJ. MAGIC, an in vivo genetic method for the rapid construction of recombinant DNA molecules. Nat Genet. 2005 Mar;37(3):311-9. DOI:10.1038/ng1505 |
MAGIC, bacterial mating approach
- Zhang Y, Werling U, and Edelmann W. SLiCE: a novel bacterial cell extract-based DNA cloning method. Nucleic Acids Res. 2012 Apr;40(8):e55. DOI:10.1093/nar/gkr1288 |
SLiCe