CH391L/S13/DNA Computing: Difference between revisions
mNo edit summary |
|||
| (45 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
[[Category:CH391L_S12]][[Category:CH391L_S13]] | [[Category:CH391L_S12]][[Category:CH391L_S13]] | ||
=Introduction= | =Introduction= | ||
DNA Computing is the performing of computations using biological molecules, and DNA specifically, instead of traditional silicon. | |||
The advantages of DNA over silicon include: | |||
* Massive parallelism: tasks can be performed simultaneously, rather than linearly in silicon computers. | |||
* Size: DNA computers are many times smaller than silicon computers. | |||
* Storage density: much, much more information can be stored in the same amount of space. | |||
The disadvantages of DNA over silicon include: | |||
* | * Impracticality: the quantities of DNA required to take advantage of its massive parallelism are unmanageable. | ||
* | * Error prone: getting biochemistry to perform as desired is still more difficult compared to silicon and electricity. | ||
* | * Speed: it still takes a long time to get your results, from start to finish, even though the computational step may be relatively rapid. | ||
The concept of using molecules in general for computation dates back to 1959 when American physicist [http://en.wikipedia.org/wiki/Richard_Feynman Richard Feynman] presented his ideas on nanotechnology, but it took until 1994 for the first successful experiment using molecules for computation to be completed. | |||
=History= | |||
[[Image:Adleman_salesman.png | "The principle of Leonard Adleman's DNA computer to solve the 'Travelling salesman' problem. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1315819/figure/f1/ Source]" | thumb|right|240px]] | |||
A computer scientist at the University of Southern California named [http://en.wikipedia.org/wiki/Leonard_Adleman Leonard Adleman] first came up with the idea of using DNA for computing purposes after reading "Molecular Biology of the Gene," a book written by James Watson (co-discoverer of the structure of DNA). | |||
In 1994 Adleman published an article in the journal Science<cite>#Adleman</cite> that proposed how DNA could be used to solve a well-known mathematical problem, called the directed [http://en.wikipedia.org/wiki/Hamiltonian_path_problem Hamilton Path problem], also known as the "traveling salesman" problem. The goal of the problem is to find the shortest path between a number of cities, going through each city only once. This experiment was significant because it showed that information could indeed be processed using the interactions between strands of synthetic DNA. | |||
''"He represented each of the seven cities as separate, singlestranded DNA molecules, 20 nucleotides long, and all possible paths between cities as DNA molecules composed of the last ten nucleotides of the departure city and the first ten nucleotides of the arrival city. Mixing the DNA strands with DNA ligase and adenosine triphosphate (ATP) resulted in the generation of all possible random paths through the cities. However, the majority of these paths were not applicable to the situation–they were either too long or too short, or they did not start or finish in the right city. Adleman then filtered out all the paths that neither started nor ended with the correct molecule and those that did not have the correct length and composition. Any remaining DNA molecules represented a solution to the problem"''<cite>#Parker</cite>. | |||
'''Here is a simpler rephrasing''' of the process of selecting only those DNA strands which were applicable: First he used PCR amplification to amplify only strands that start and end at the correct cities, A and G. Then he used gel electrophoresis to filter out all the strands which were not of the correct length (i.e. strands which visited each of seven towns only once would only contain links between six towns). Finally, he used affinity purification to filter out strands which do not visit each town (i.e. any strands which do not contain town A are first discarded, then those which do not contain B, and so on). Any strands left over must represent a valid route. In this case, the only valid route was encoded by the strand ABBCCDDEEFFG: A to B to C to D to E to F to G. | |||
=Challenges= | |||
Adleman's experiment is an example of an [http://en.wikipedia.org/wiki/NP-complete NP-complete problem] (from non-polynomial) for which no known efficient (i.e. polynomial time) algorithm exists. It has been proposed that these types of problems are ideal for the massive parallelism of biology. Whereas traditional computers are designed to perform one calculation very fast, DNA strands can produce billions of potential answers simultaneously. | |||
However, because in most difficult problems the number of possible solutions grows exponentially with the size of the problem, even relatively small problems would require massive amounts of DNA to represent all possible answers. For example, it has been estimated that scaling up the Hamilton Path Problem to 200 cities would require a weight of DNA that exceeds that of the earth<cite>#NCBI</cite>. For this reason and others, the field has changed focus to other areas. | |||
=DNA-based Logic= | |||
[[Image:Strand_displacement.png | A toehold-mediated strand displacement reaction. The red strand binds to the green strand, displacing and freeing the blue strand. [http://en.wikipedia.org/wiki/File:Strand_displacement.svg source] | thumb|right|240px]] | |||
The fundamental building blocks of computing are based on logic circuits or "gates", such as AND, OR, NOT. Using Adleman's work, other researchers have been developing DNA-based logic circuits that are useful in a variety of ways, like performing mathematical and logical operations and recognizing patterns. Progress is even being made in in-cell programming, opening up the doors to, for example, advanced cancer treatments. | |||
==Strand Displacement== | |||
Perhaps the best-known advances in the field are by a group of researchers at Caltech led by Erik Winfree. In strand-displacement, the inputs are free-floating single strands of DNA or RNA, and [http://en.wikipedia.org/wiki/Logic_gate logic gates] are made from two or more strands, with one being the future output signal. If an input strand has a complementary "tab" to a logic gate, it binds and then displaces the output strand, which then detaches. The output strand then becomes an input to another logic gate, in propagation. In a 2011 paper, Winfree together with colleague Lulu Qian used this technique to build a circuit, made of 74 different DNA strands, capable of calculating the square roots of four-digit binary numbers (aka 0 to 15)<cite>#Seelig</cite>. | |||
== | ==DNAzymes== | ||
At Columbia University, Milan Stojanovic and his group have built circuits using a sister to strand displacement based on deoxyribozymes or DNAzymes (synthetically made, single-stranded DNA sequences that cut other DNA strands in specific places). They make logic gates by adding "stem loops" to the DNAzyme, which prevent it from working. When input strands bind to complementary sequences on the loop, the loop breaks, thus allowing the DNAzyme to activate and turning the gate on. It then interacts with other strands in a cascading fashion, allowing the creation of more complex logic. Stojanovic's group used this method to build circuits able to play tic-tac-toe<cite>#Stojanovic</cite>. | |||
== | ==Enzymes== | ||
Logic can also be created using methods that more closely resemble natural operations inside of living cells. A group led by Yaakov Benenson at the Swiss Federal Institute of Technology (ETH Zurich) and Ron Weiss of MIT is creating circuits using common enzymes like polymerases, nucleases and exonucleases that operate inside cells, utilizing existing cellular infrastructure. In 2011, the team created a circuit capable of first recognizing cervical cancer and then destroying the cell it is found within. It works by looking for five microRNAs specific to cervical cancer; when all five are at the right levels, the circuit activates, producing a protein that causes the cell to self-destruct<cite>#Xie</cite>. | |||
==Algorithmic self-assembly== | |||
[[Image:Sierpinski_gasket.png | DNA arrays that display a representation of the Sierpinski gasket on their surfaces. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC534809/ source] | thumb|right|240px]] | |||
DNA computing has benefitted from the separate and distinct field of [http://en.wikipedia.org/wiki/DNA_nanotechnology DNA nanotechnology]. | |||
The founder of the field, American biochemist Nadrian Seeman, has worked with Erik Winfree to show how two-dimensional sheets of DNA "tiles" could self assemble into other, larger structures. Winfree and his student Paul Rothemund then demonstrated how these structures could be used for computing purposes in a 2004 paper that describes the self assembly of a fractal called a Sierpinski gasket, which won them the 2006 Feynman Prize in Nanotechnology. Winfree's major insight was that the tiles could be used as "Wang tiles", meaning the assembly of the structure was capable of computation<cite>#Rothemund</cite>. | |||
=Storage= | |||
[[Image:Church_book.png | Church's methodology for encoding and decoding 700 terabytes of information in DNA. [http://www.extremetech.com/wp-content/uploads/2012/08/coding-decoding-dna-storage.jpg source] | thumb|right|240px]] | |||
DNA can be treated as a highly stable and compact storage medium. In 2012, George Church, a bioengineer and geneticist at Harvard, succeeded in storing 1000 times the largest data size previously achieved using DNA. Church encoded 700 terabytes of information into a single gram of DNA. For comparison, storing the same amount of information using the densest electronic medium would require 330 pounds of hard-drives. | |||
His methodology was synthesizing [http://en.wikipedia.org/wiki/Word_(computer_architecture) 96-bit] strands, with each base representing a binary value (ex: T and G = 1, A and C = 0). The "data" in question was 70 billion copies of his latest book, [http://www.perseusbooksgroup.com/basic/book_detail.jsp?isbn=0465021751 "Regenesis: How Synthetic Biology Will Reinvent Nature and Ourselves"] | |||
To read the DNA, Church simply sequenced it using next-generation sequencing, converting each of the bases back into binary. Each strand of DNA had a 19-bit address block at the start, so the DNA can be sequenced out of order and then sorted back into usable data by following the addresses. | |||
= | =iGEM Connection= | ||
[[Image:BIL_gates.png | Drew Endy's "Boolean Integrase Logic" (BIL) gates. [http://youtu.be/ahYZBeP_r5U source] | thumb|right|240px]] | |||
On March 28, 2013, Drew Endy and his team of researchers at Stanford University published a paper<cite>#Bonnet</cite> in Science that outlines how to create single-layer, transcriptor-based logic gates utilizing a three-terminal device architecture to control the flow of RNA polymerase along DNA. Endy has open-sourced these transcriptors through the Registry of Standard Biological Parts, and an informational video is available [http://youtu.be/ahYZBeP_r5U here]. | |||
A complementary development was published in Nature (January 2013) by scientist Timothy Lu at MIT that details how to store information about a cell's lifecycle events, like key moments in a cell's ancestry<cite>#Siuti</cite>. | |||
There is a great animated infographic at the end of [http://www.npr.org/2013/03/29/175604770/tiny-dna-switches-aim-to-revolutionize-cellular-computing. this article] that helps explain the concepts. | |||
=Future Directions= | |||
The most popular narrative surrounding the future of DNA computing and DNA-based logic is that one day the technology could be used to program living cells inside our bodies. For example, they could "detect disease, warn of toxic threats and, where danger lurked, even self-destruct cells gone rogue"<cite>#MercuryNews</cite>. To quote Drew Endy: "We're going to be able to put computers inside any living cell you want... Any place you want a little bit of logic, a little bit of computation, a little bit of memory -- we're going to be able to do that"<cite>#MercuryNews</cite>. Whether or not this prediction will come to fruition is up for debate, as the technology has its detractors. For example, many who participated in this year's synthetic biology class (Spring 2013) believed that natural evolutionary processes will always confound the efforts of those trying to bound the behavior of a living cell in vivo. However, no published critiques in the public domain could be found at the time of this writing. | |||
==References== | ==References== | ||
<biblio> | <biblio> | ||
# | #Adleman [http://www.usc.edu/dept/molecular-science/papers/fp-sci94.pdf Adleman, L. M. (1994). "Molecular computation of solutions to combinatorial problems". Science 266 (5187): 1021–1024. doi:10.1126/science.7973651. PMID 7973651.] | ||
# | #Parker [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1315819/ Parker, J. "Computing with DNA", EMBO Rep. 2003 Jan;4(1):7-10., PMID: 12524509] | ||
# | #Seelig [http://www.sciencemag.org/content/314/5805/1585 Seelig, G.; Soloveichik, D.; Zhang, D. Y.; Winfree, E. (8 December 2006). "Enzyme-free nucleic acid logic circuits". Science 314 (5805): 1585–1588. doi:10.1126/science.1132493. PMID 17158324] | ||
#Stojanovic [http://pubs.acs.org/doi/full/10.1021/ja016756v# Milan N. Stojanovic, Tiffany Elizabeth Mitchell, and Darko Stefanovic (2002) Deoxyribozyme-Based Logic Gates Journal of the American Chemical Society 124(14), 3555-3561] | |||
# | #Xie [http://www.sciencemag.org/content/333/6047/1307nature02551.html Xie, Z et al., "Multi-Input RNAi-Based Logic Circuit for Identification of Specific Cancer Cells", Science 2 September 2011: Vol. 333 no. 6047 pp. 1307-1311 DOI: 10.1126/science.1205527] | ||
# | #Rothemund [http://www.plosbiology.org/article/info:doi/10.1371/journal.pbio.0020424 Rothemund, P. W. K.; Papadakis, N.; Winfree, E. (2004). "Algorithmic Self-Assembly of DNA Sierpinski Triangles". PLoS Biology 2 (12): e424. doi:10.1371/journal.pbio.0020424. PMC 534809. PMID 15583715] | ||
# | #Bonnet [http://www.sciencemag.org/content/early/2013/03/27/science.1232758 Bonnet et. al., "Amplifying Genetic Logic Gates", Published Online March 28 2013, Science DOI: 10.1126/science.1232758] | ||
# | #Siuti [http://www.nature.com/nbt/journal/vaop/ncurrent/full/nbt.2510.html Siuti, P., Yazbek, J. & Lu, T. K. Nature Biotech. advance online publication, http://dx.doi.org/10.1038/nbt.2510 (2013)] | ||
#NCBI [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1315819/ http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1315819/] | |||
#MercuryNews [http://www.mercurynews.com/science/ci_22891433/biological-computer-created-at-stanford http://www.mercurynews.com/science/ci_22891433/biological-computer-created-at-stanford] | |||
</biblio> | </biblio> | ||
Latest revision as of 14:42, 2 May 2013
Introduction
DNA Computing is the performing of computations using biological molecules, and DNA specifically, instead of traditional silicon.
The advantages of DNA over silicon include:
- Massive parallelism: tasks can be performed simultaneously, rather than linearly in silicon computers.
- Size: DNA computers are many times smaller than silicon computers.
- Storage density: much, much more information can be stored in the same amount of space.
The disadvantages of DNA over silicon include:
- Impracticality: the quantities of DNA required to take advantage of its massive parallelism are unmanageable.
- Error prone: getting biochemistry to perform as desired is still more difficult compared to silicon and electricity.
- Speed: it still takes a long time to get your results, from start to finish, even though the computational step may be relatively rapid.
The concept of using molecules in general for computation dates back to 1959 when American physicist Richard Feynman presented his ideas on nanotechnology, but it took until 1994 for the first successful experiment using molecules for computation to be completed.
History

A computer scientist at the University of Southern California named Leonard Adleman first came up with the idea of using DNA for computing purposes after reading "Molecular Biology of the Gene," a book written by James Watson (co-discoverer of the structure of DNA).
In 1994 Adleman published an article in the journal Science[1] that proposed how DNA could be used to solve a well-known mathematical problem, called the directed Hamilton Path problem, also known as the "traveling salesman" problem. The goal of the problem is to find the shortest path between a number of cities, going through each city only once. This experiment was significant because it showed that information could indeed be processed using the interactions between strands of synthetic DNA.
"He represented each of the seven cities as separate, singlestranded DNA molecules, 20 nucleotides long, and all possible paths between cities as DNA molecules composed of the last ten nucleotides of the departure city and the first ten nucleotides of the arrival city. Mixing the DNA strands with DNA ligase and adenosine triphosphate (ATP) resulted in the generation of all possible random paths through the cities. However, the majority of these paths were not applicable to the situation–they were either too long or too short, or they did not start or finish in the right city. Adleman then filtered out all the paths that neither started nor ended with the correct molecule and those that did not have the correct length and composition. Any remaining DNA molecules represented a solution to the problem"[2].
Here is a simpler rephrasing of the process of selecting only those DNA strands which were applicable: First he used PCR amplification to amplify only strands that start and end at the correct cities, A and G. Then he used gel electrophoresis to filter out all the strands which were not of the correct length (i.e. strands which visited each of seven towns only once would only contain links between six towns). Finally, he used affinity purification to filter out strands which do not visit each town (i.e. any strands which do not contain town A are first discarded, then those which do not contain B, and so on). Any strands left over must represent a valid route. In this case, the only valid route was encoded by the strand ABBCCDDEEFFG: A to B to C to D to E to F to G.
Challenges
Adleman's experiment is an example of an NP-complete problem (from non-polynomial) for which no known efficient (i.e. polynomial time) algorithm exists. It has been proposed that these types of problems are ideal for the massive parallelism of biology. Whereas traditional computers are designed to perform one calculation very fast, DNA strands can produce billions of potential answers simultaneously.
However, because in most difficult problems the number of possible solutions grows exponentially with the size of the problem, even relatively small problems would require massive amounts of DNA to represent all possible answers. For example, it has been estimated that scaling up the Hamilton Path Problem to 200 cities would require a weight of DNA that exceeds that of the earth[3]. For this reason and others, the field has changed focus to other areas.
DNA-based Logic

The fundamental building blocks of computing are based on logic circuits or "gates", such as AND, OR, NOT. Using Adleman's work, other researchers have been developing DNA-based logic circuits that are useful in a variety of ways, like performing mathematical and logical operations and recognizing patterns. Progress is even being made in in-cell programming, opening up the doors to, for example, advanced cancer treatments.
Strand Displacement
Perhaps the best-known advances in the field are by a group of researchers at Caltech led by Erik Winfree. In strand-displacement, the inputs are free-floating single strands of DNA or RNA, and logic gates are made from two or more strands, with one being the future output signal. If an input strand has a complementary "tab" to a logic gate, it binds and then displaces the output strand, which then detaches. The output strand then becomes an input to another logic gate, in propagation. In a 2011 paper, Winfree together with colleague Lulu Qian used this technique to build a circuit, made of 74 different DNA strands, capable of calculating the square roots of four-digit binary numbers (aka 0 to 15)[4].
DNAzymes
At Columbia University, Milan Stojanovic and his group have built circuits using a sister to strand displacement based on deoxyribozymes or DNAzymes (synthetically made, single-stranded DNA sequences that cut other DNA strands in specific places). They make logic gates by adding "stem loops" to the DNAzyme, which prevent it from working. When input strands bind to complementary sequences on the loop, the loop breaks, thus allowing the DNAzyme to activate and turning the gate on. It then interacts with other strands in a cascading fashion, allowing the creation of more complex logic. Stojanovic's group used this method to build circuits able to play tic-tac-toe[5].
Enzymes
Logic can also be created using methods that more closely resemble natural operations inside of living cells. A group led by Yaakov Benenson at the Swiss Federal Institute of Technology (ETH Zurich) and Ron Weiss of MIT is creating circuits using common enzymes like polymerases, nucleases and exonucleases that operate inside cells, utilizing existing cellular infrastructure. In 2011, the team created a circuit capable of first recognizing cervical cancer and then destroying the cell it is found within. It works by looking for five microRNAs specific to cervical cancer; when all five are at the right levels, the circuit activates, producing a protein that causes the cell to self-destruct[6].
Algorithmic self-assembly

DNA computing has benefitted from the separate and distinct field of DNA nanotechnology. The founder of the field, American biochemist Nadrian Seeman, has worked with Erik Winfree to show how two-dimensional sheets of DNA "tiles" could self assemble into other, larger structures. Winfree and his student Paul Rothemund then demonstrated how these structures could be used for computing purposes in a 2004 paper that describes the self assembly of a fractal called a Sierpinski gasket, which won them the 2006 Feynman Prize in Nanotechnology. Winfree's major insight was that the tiles could be used as "Wang tiles", meaning the assembly of the structure was capable of computation[7].
Storage

DNA can be treated as a highly stable and compact storage medium. In 2012, George Church, a bioengineer and geneticist at Harvard, succeeded in storing 1000 times the largest data size previously achieved using DNA. Church encoded 700 terabytes of information into a single gram of DNA. For comparison, storing the same amount of information using the densest electronic medium would require 330 pounds of hard-drives.
His methodology was synthesizing 96-bit strands, with each base representing a binary value (ex: T and G = 1, A and C = 0). The "data" in question was 70 billion copies of his latest book, "Regenesis: How Synthetic Biology Will Reinvent Nature and Ourselves"
To read the DNA, Church simply sequenced it using next-generation sequencing, converting each of the bases back into binary. Each strand of DNA had a 19-bit address block at the start, so the DNA can be sequenced out of order and then sorted back into usable data by following the addresses.
iGEM Connection
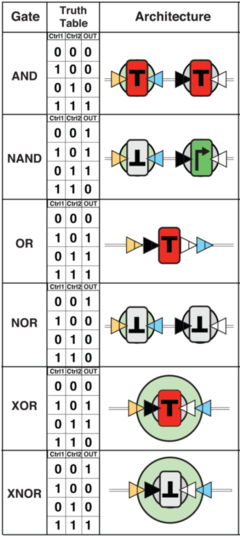
On March 28, 2013, Drew Endy and his team of researchers at Stanford University published a paper[8] in Science that outlines how to create single-layer, transcriptor-based logic gates utilizing a three-terminal device architecture to control the flow of RNA polymerase along DNA. Endy has open-sourced these transcriptors through the Registry of Standard Biological Parts, and an informational video is available here.
A complementary development was published in Nature (January 2013) by scientist Timothy Lu at MIT that details how to store information about a cell's lifecycle events, like key moments in a cell's ancestry[9].
There is a great animated infographic at the end of this article that helps explain the concepts.
Future Directions
The most popular narrative surrounding the future of DNA computing and DNA-based logic is that one day the technology could be used to program living cells inside our bodies. For example, they could "detect disease, warn of toxic threats and, where danger lurked, even self-destruct cells gone rogue"[10]. To quote Drew Endy: "We're going to be able to put computers inside any living cell you want... Any place you want a little bit of logic, a little bit of computation, a little bit of memory -- we're going to be able to do that"[10]. Whether or not this prediction will come to fruition is up for debate, as the technology has its detractors. For example, many who participated in this year's synthetic biology class (Spring 2013) believed that natural evolutionary processes will always confound the efforts of those trying to bound the behavior of a living cell in vivo. However, no published critiques in the public domain could be found at the time of this writing.