CH391L/S13/CAD
From OpenWetWare
Representing Synthetic Biology Designs
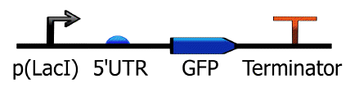
- The Synthetic Biology Open Language is an open-source standard for representing designs consisting of both DNA sequence information and higher level annotation of parts with defined roles and behaviors [1]. The core specification of this system has been developed as an RFC [2]. Several different synthetic biology CAD software programs use this format. Representation at this higher level of parts can be visualized and simulated in some of these systems (e.g., TinkerCell).

- The Eugene Language is an open-source human-readable language designed to facilitate automatic creation of new devices from a collection of parts. Eugene includes a standardized format for specifying devices and parts as well as constraints on how they can be assembled into higher level devices (i.e. genetic toggle switch). Eugene also features functions for automatic generation of functional assemblies into complex devices.
References
- Galdzicki M, Rodriguez C, Chandran D, Sauro HM, and Gennari JH. Standard biological parts knowledgebase. PLoS One. 2011 Feb 24;6(2):e17005. DOI:10.1371/journal.pone.0017005 |
Standard biological parts knowledgebase
-
Synthetic Biology Open Language (SBOL) Version 1.0.0