CH391L/S13/CAD: Difference between revisions
No edit summary |
No edit summary |
||
| Line 4: | Line 4: | ||
The [http://www.sbolstandard.org Synthetic Biology Open Language] is an open-source standard for representing designs consisting of both DNA sequence information and higher level annotation of parts with defined roles and behaviors <cite>Galdzicki2011</cite>. The core specification of this system has been developed as an RFC <cite>SBOLRFC</cite>. Several different [http://www.sbolstandard.org/sbolstandard/software-tools-using-sbol synthetic biology CAD software programs] use this format. Representation at this higher level of parts can be visualized and simulated in some of these systems (e.g., [http://www.tinkercell.com/ TinkerCell]). | The [http://www.sbolstandard.org Synthetic Biology Open Language] is an open-source standard for representing designs consisting of both DNA sequence information and higher level annotation of parts with defined roles and behaviors <cite>Galdzicki2011</cite>. The core specification of this system has been developed as an RFC <cite>SBOLRFC</cite>. Several different [http://www.sbolstandard.org/sbolstandard/software-tools-using-sbol synthetic biology CAD software programs] use this format. Representation at this higher level of parts can be visualized and simulated in some of these systems (e.g., [http://www.tinkercell.com/ TinkerCell]). | ||
The [http://www.eugenecad.org/ Eugene Language] is an open-source human-readable language designed to facilitate automatic creation of new devices from a collection of parts. Eugene includes a standardized format for specifying devices and parts as well as constraints on how they can be assembled into higher level devices (i.e. genetic toggle switch). Eugene also features functions for automatic generation of functional assemblies into complex devices. Eugene does not support visualization of constructs. | The [http://www.eugenecad.org/ Eugene Language]<cite>Eugene2011</cite> is an open-source human-readable language designed to facilitate automatic creation of new devices from a collection of parts. Eugene includes a standardized format for specifying devices and parts as well as constraints on how they can be assembled into higher level devices (i.e. genetic toggle switch). Eugene also features functions for automatic generation of functional assemblies into complex devices. Eugene does not support visualization of constructs. | ||
==Synthetic Biology CAD Tools == | ==Synthetic Biology CAD Tools == | ||
| Line 33: | Line 33: | ||
#VectorEditor2012 pmid=22718978 | #VectorEditor2012 pmid=22718978 | ||
//Design, implementation and practice of JBEI-ICE: an open source biological part registry platform and tools. | //Design, implementation and practice of JBEI-ICE: an open source biological part registry platform and tools. | ||
#Eugene2011 pmid=21559524 | |||
//Eugene--a domain specific language for specifying and constraining synthetic biological parts, devices, and systems | |||
</biblio> | </biblio> | ||
Revision as of 02:11, 3 February 2013
Representing Synthetic Biology Designs
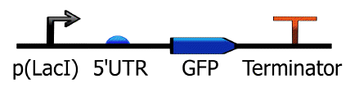
The Synthetic Biology Open Language is an open-source standard for representing designs consisting of both DNA sequence information and higher level annotation of parts with defined roles and behaviors [1]. The core specification of this system has been developed as an RFC [2]. Several different synthetic biology CAD software programs use this format. Representation at this higher level of parts can be visualized and simulated in some of these systems (e.g., TinkerCell).
The Eugene Language[3] is an open-source human-readable language designed to facilitate automatic creation of new devices from a collection of parts. Eugene includes a standardized format for specifying devices and parts as well as constraints on how they can be assembled into higher level devices (i.e. genetic toggle switch). Eugene also features functions for automatic generation of functional assemblies into complex devices. Eugene does not support visualization of constructs.
Synthetic Biology CAD Tools
Synthetic Biology Computer Assisted Design (CAD) tools are programs which help to create novel biological constructs. At the most basic, these programs are essentially enhanced DNA editors which provide a user interface to facilitate easier manipulation of the basic “parts” which comprise biological devices. Some of the more advanced programs have a variety of functions including visualization, asserting validity of constructs, and simulations of metabolic networks.
Simulation Software
Design Tools
References
- Galdzicki M, Rodriguez C, Chandran D, Sauro HM, and Gennari JH. Standard biological parts knowledgebase. PLoS One. 2011 Feb 24;6(2):e17005. DOI:10.1371/journal.pone.0017005 |
Standard biological parts knowledgebase
-
Synthetic Biology Open Language (SBOL) Version 1.0.0
- Bilitchenko L, Liu A, Cheung S, Weeding E, Xia B, Leguia M, Anderson JC, and Densmore D. Eugene--a domain specific language for specifying and constraining synthetic biological parts, devices, and systems. PLoS One. 2011 Apr 29;6(4):e18882. DOI:10.1371/journal.pone.0018882 |
Eugene--a domain specific language for specifying and constraining synthetic biological parts, devices, and systems
- Chandran D, Bergmann FT, and Sauro HM. TinkerCell: modular CAD tool for synthetic biology. J Biol Eng. 2009 Oct 29;3:19. DOI:10.1186/1754-1611-3-19 |
TinkerCell: modular CAD tool for synthetic biology
- Weeding E, Houle J, and Kaznessis YN. SynBioSS designer: a web-based tool for the automated generation of kinetic models for synthetic biological constructs. Brief Bioinform. 2010 Jul;11(4):394-402. DOI:10.1093/bib/bbq002 |
SynBioSS designer: a web-based tool for the automated generation of kinetic models for synthetic biological constructs
- Ham TS, Dmytriv Z, Plahar H, Chen J, Hillson NJ, and Keasling JD. Design, implementation and practice of JBEI-ICE: an open source biological part registry platform and tools. Nucleic Acids Res. 2012 Oct;40(18):e141. DOI:10.1093/nar/gks531 |
Design, implementation and practice of JBEI-ICE: an open source biological part registry platform and tools.
- Villalobos A, Ness JE, Gustafsson C, Minshull J, and Govindarajan S. Gene Designer: a synthetic biology tool for constructing artificial DNA segments. BMC Bioinformatics. 2006 Jun 6;7:285. DOI:10.1186/1471-2105-7-285 |
Gene Designer:a synthetic biology tool for constructing artificial DNA segments