CH391L/S12/GeneandGenomeSynthesis
Gene and Genome Synthesis
Introduction
Gene synthesis, or artificial gene synthesis, refers to the process of creating a nucleic acid template for a gene in vitro, without the requirement of a preexisting DNA template. Soon after the elucidation of the genetic code and the description of the central dogma of molecular biology, there arose a need to synthesize genes de novo in order to study their biological function both in the test tube and in model organisms. Chemical synthesis of DNA has grown from an expensive and time-consuming process into a viable commercial industry capable of high-throughput manufacture of almost any scale of custom DNA molecules in almost any context. This allows species-specific gene optimization, creation of genes from rare or dangerous sources, and combinatorial assembly of any DNA sequence that can be chemically synthesized, even including non-traditional bases. The most advanced applications of gene synthesis have been applied to the recent creation of completely synthetic minimal genomes in prokaryotes.
Despite nearly four decades of progress in gene synthesis technologies, most DNA sequences used in modern molecular biology are assembled in part or in whole from naturally occurring templates. However this limits the scope and applications to previously existing genes and the results of large-scale genomic surveys of novel genes from nature. Modern gene synthesis relies heavily on advancements in chemical DNA oligonucleotide synthesis, with the primary challenges being scale, cost, fidelity and the eventual assembly of complete gene products.
History of Gene Synthesis
Gene synthesis predates the invention of restriction enzymes and molecular cloning techniques by several years. The first gene to be completely synthesized in vitro was a 77-nt alanine transfer RNA by the laboratory of Har Gobind Khorana in 1972. This was the result of nearly five years of work and resulted in a DNA template without promoter or transcriptional control sequences. The first peptide- and protein-producing synthetic genes were created in 1977 and 1979, respectively (Itakura 1977, Goeddell 1979). Steady advancement has led to recent synthesis of complete gene clusters tens of thousands of nucleotides in length, and ultimately a bacterial genome approximately 1.2 million bases in length.
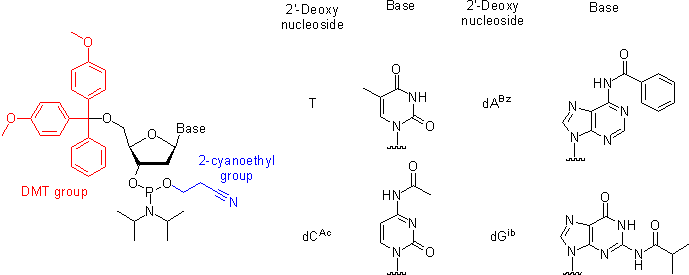
Oligonucleotide Synthesis
Regardless of the length of the eventual product, synthetic DNA constructs are built from some combination of short DNA oligonucleotides. These oligonucleotides are later assembled into a complementary DNA duplex, amplified and inserted into their final genetic context.
Oligonucleotides are chemically synthesized from DNA phosphoramidite monomers. Briefly, activated phosphoramidite monomers are added in the 3' to 5' direction using a cyclical activation and blocking chemistry to obtain a DNA polymer linked by phosphodiester bonds.
Summary of Methods
Ligation-Based Methods
In ligation-mediated assembly, the synthetic gene or DNA fragment of interest is broken up into individual overlapping DNA oligonucleotides that cover both strands of the eventual DNA duplex. In contrast to PCR-based methods of gene synthesis, there are no gaps introduced in either strand during design, but rather the oligonucleotides completely reconstitute the eventual DNA target.
The oligonucleotides are chemically synthesized and phosphorylated at their 5' ends using purified kinase in an in vitro reaction. After complementary oligo fragments are annealed using using thermal cycling, purified DNA ligase is added to the reaction in order to splice together the 3' OH and 5' phosphate gaps on each backbone.
PCR-based Methods
PCA
Solid-Phase Synthesis
Blue Heron
Multiplex/Microchip Synthesis
Oligos are commodity, still expensive on large scale Microarray based oligo synth - Inkjets, photolithography, electrochemical Chips allow large numbers of oligos, small scale. On-chip amplification with universal PCR primers in frags can amplify pools to levels useful in gene synthesis methods. Oligo error rates reduced in multiplex using hybridization screening arrays to eliminate mismatched oligos. Nonselected oligo synthesis methods have error rate of 1 in ~160 bp, selected multiplex oligo methods have 1 in ~1,500 error rate. Further work could extend this to 1 in 10^6 using mismatch proteins. Multiplex uses: ITT protein arrays, synthesis of operons (Tian 2004 ribosomal operon from E. coli, 21 genes and codon optimization)