Biomolecular Breadboards:Preliminary Data: Difference between revisions
Clare Hayes (talk | contribs) mNo edit summary |
|||
| (28 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{{Template:Biomolecular Breadboards}} | {{Template:Biomolecular Breadboards}} | ||
= | This page contains some data that we have taken with the TX-TL breadboard. | ||
{| style="float: right" | |||
|- | |||
| __TOC__ | |||
|} | |||
==Plasmid Expression of GFP== | |||
Using pBEST-OR2-OR1-Pr-UTR1- | Using pBEST-OR2-OR1-Pr-UTR1-deGFP-T500, a plasmid enhanced for GFP expression, the biomolecular breadboard is able to express mass at equal concentrations to comparable bacteriophage in-vitro systems (J. Shin and V. Noireaux, 2010). | ||
Expression of plasmids can be optimized by concentration. | Expression of plasmids can be optimized by concentration. Figure 1 (below) shows a typical time response for expression of the control plasmid, along with the data we collected from our plate reader. If you are trying to get TX-TL working, this is probably the first test that you should do. | ||
{| border=1 width=100% | |||
|- | |||
| | |||
{| | |||
|- | |||
| | |||
[[Image:plasmid_sat.png|400px]] | [[Image:plasmid_sat.png|400px]] | ||
| valign=top | | |||
* Raw data file: [[Media:plasmid_deGFP-070812.xls|plasmid_deGFP-070812.xls]] (Excel spreadsheet) | |||
* Information about protocol on "Outline" sheet; plotted data is on the sheet "plotted-data" | |||
<hr> | |||
* Control plasmid file: [[Media:pBEST_OR2_OR1_Pr_UTR-deGFP-T500.gb|pBEST_OR2_OR1_Pr_UTR-deGFP-T500.gb]] (GenBank format) | |||
* More information: [[Biomolecular Breadboards:pBEST_OR2_OR1_Pr_UTR-deGFP-T500.gb|Control plasmid description]] | |||
|} | |||
'''Figure 1. eGFP expression as a function of plasmid DNA template.''' Plasmid DNA pBEST-OR2-OR1-Pr-UTR1-eGFP-T500 is varied by concentration. | |||
|} | |||
==Protecting Linear DNA from Exonuclease-Mediated Degradation== | |||
Current standards for circuit design utilize plasmids for DNA template, which require time-consuming subcloning steps. However, circuits based on linear DNA require only PCR assembly or gene synthesis, which drastically decreases preparation time. As a purely extract-derived system, our biomolecular breadboard exhibits exonuclease activity which degrades linear DNA. We are developing multiple technologies to protect linear DNA from exonuclease degradation. These include: | Current standards for circuit design utilize plasmids for DNA template, which require time-consuming subcloning steps. However, circuits based on linear DNA require only PCR assembly or gene synthesis, which drastically decreases preparation time. As a purely extract-derived system, our biomolecular breadboard exhibits exonuclease activity which degrades linear DNA. We are developing multiple technologies to protect linear DNA from exonuclease degradation. These include: | ||
# Protecting linear DNA using noncoding segments | # Protecting linear DNA using noncoding segments | ||
| Line 17: | Line 34: | ||
# Adding thiosulfate bonds to 5' ends | # Adding thiosulfate bonds to 5' ends | ||
===Protection Sequences=== | |||
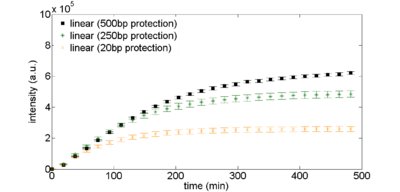
The simplest way to protect linear DNA is to put additional "junk" DNA around the construct you are testing. Figure 2 shows the expression curves for different lengths of protection sequences. The overall expression with long protection sequences is about 2X less than what you see with plasmid DNA (compare the plots below to the 2nM plasmid DNA expression levels in Figure 1). | |||
{| border=1 width=100% | |||
|- | |||
| | |||
[[Image:linear.png|400px]] | [[Image:linear.png|400px]] | ||
<br>'''Figure 2: Exonuclease protection using non-coding DNA.''' Linear DNA templates, 2nM, are derived from plasmid DNA pBEST-OR2-OR1-Pr-UTR1-eGFP-Del6-229-T500 with varying amounts of noncoding DNA surrounding the coding sequence. | <br>'''Figure 2: Exonuclease protection using non-coding DNA.''' Linear DNA templates, 2nM, are derived from plasmid DNA pBEST-OR2-OR1-Pr-UTR1-eGFP-Del6-229-T500 with varying amounts of noncoding DNA surrounding the coding sequence. | ||
|} | |||
===GamS=== | |||
The GamS protein can be used to protect linear DNA. GamS is a shortened version of the lambda phage Gam protein, which inhibits activity of the exonuclease RecBCD ([http://www.ncbi.nlm.nih.gov/pubmed/17583735 Murphy, 2007]). Figures 3 and 4 show time responses for expression of linear DNA using GamS. Using a combination of GamS and protection sequences, it is possible to get approximately 60% of the expression that you get from plasmid DNA. | |||
{| border=1 width=100% | |||
|- | |||
| | |||
[[Image:gamSComp.png|400px]] | [[Image:gamSComp.png|400px]] | ||
<br>'''Figure 3. GamS from lambda phage, an inhibitor of RecBCD complex, inhibits template DNA degradation.''' Linear DNA templates, 2nM, are derived from plasmid DNA pBEST-OR2-OR1-Pr-UTR1-eGFP-Del6-229-T500. GamS supplied at 3uM concentration. | [[Image:gamS-protsynt81bis_05Sep12.png|400px]] | ||
<br>'''Figure 3. (left) GamS from lambda phage, an inhibitor of RecBCD complex, inhibits template DNA degradation.''' Linear DNA templates, 2nM, are derived from plasmid DNA pBEST-OR2-OR1-Pr-UTR1-eGFP-Del6-229-T500. GamS supplied at 3uM concentration. (right) Simulation results based on a simple ODE model. | |||
|} | |||
{| border=1 width=100% | |||
|- | |||
| | |||
[[Image:linear_sat.png|400px]] | [[Image:linear_sat.png|400px]] | ||
<br>'''Figure 4. eGFP expression as a function of linear DNA template, with gamS.''' Plasmid DNA pBEST-OR2-OR1-Pr-UTR1-eGFP-Del6-229-T500 is varied by concentration. GamS supplied at 3uM concentration. | <br>'''Figure 4. eGFP expression as a function of linear DNA template, with gamS.''' Plasmid DNA pBEST-OR2-OR1-Pr-UTR1-eGFP-Del6-229-T500 is varied by concentration. GamS supplied at 3uM concentration. | ||
|} | |||
Note that you can include GamS in your reactions either by putting in the DNA encoding GamS or by supplying purified GamS into the reaction. We recommend the latter since using DNA to express GamS uses up some of the resources that would otherwise power your circuit, and also takes a while before you express enough GamS to protect linear DNA. | |||
===Thiosulfate Bonds=== | |||
We have also tried using thiosulfate bonds to protect linear DNA. These have little effect. | |||
{| border=1 width=100% | |||
|- | |||
| | |||
[[Image:ts-1.png|400px]] | [[Image:ts-1.png|400px]] | ||
<br>'''Figure 5. Thiosulfate bonds protect against 5' exonuclease degradation.''' Linear DNA templates, 2nM, are derived from plasmid DNA pBEST-OR2-OR1-Pr-UTR1-eGFP-Del6-229-T500. 5 thiosulfate bonds are present at each 5' end. GamS supplied at 3uM concentration. | <br>'''Figure 5. Thiosulfate bonds protect against 5' exonuclease degradation.''' Linear DNA templates, 2nM, are derived from plasmid DNA pBEST-OR2-OR1-Pr-UTR1-eGFP-Del6-229-T500. 5 thiosulfate bonds are present at each 5' end. GamS supplied at 3uM concentration. | ||
|} | |||
== Protein Degradation == | |||
Implementing protein degradation is integral to enabling the function of many dynamical circuits, such as oscillators or feed-forward loops. We have been exploring the overexpression of AAA ATPases ClpXP to selectively target proteins with degradation tags. Initial experiments indicate that ClpXP, when overexpressed, can accelerate degradation of purified eGFP-ssrA (Fig. 1). We will further characterize this AAA ATPase family, as well as try to demonstrate its use through an incoherent feed-forward loop. We have not been able to replicate the results by adding purified ClpXP protein. However, we intend to express ClpXP off of plasmids or to create custom extract with ClpXP already overexpressed to enable rapid protein degradation. | |||
{| border=1 width=100% | |||
|- | |||
| | |||
[[Image:txtl-clipxp.png|400px]] | |||
<br>'''Figure 6. Protein degradation can be increased using endogenous pathways.''' ClpX and ClpP are AAA proteases which selectively target proteins with proteolytic tags. 763ng of purified eGFP-ssrA is degraded over time in either extract with endogenous amounts of ClpX and ClpP (“control”), or ClpX and ClpP upregulated in a 6:1 ratio (“over-expression”). ClpX and ClpP are encoded on plasmids under an IPTG-inducible T7 promoter. Reaction also incubated with 0.2nM of plasmid encoding for sigma70-driven T7 RNA polymerase. Up to 6-fold increase in degradation rate can be seen. | |||
|} | |||
== Environmental Context == | |||
The TX-TL system can be used to study the operation of circuits in different environmental contexts, including salts, energy levels and temperature. | |||
=== Magnesium and Potassium concentration === | |||
{| border=1 width=100% | |||
|- | |||
| | |||
<center> | |||
[[Image:txtl-mgk-cascade-circuit.png|400px]] <br> | |||
[[Image:txtl-mgk-cascade-data.png|600px]] | |||
</center> | |||
<br>'''Figure 7: Response of two two-stage transcriptional activation cascades as a function of magnesium and potassium.''' Top: schematic of the circuit where x and y are either σ19 or σ38. Bottom: response of the two circuits as a function of magnesium (A) and potassium (B). | |||
|} | |||
=== Toxicity === | |||
To optimize TX-TL for the addition of proteins of interest or reagents (such as gamS for linear protection and ClpX for protein degradation), a toxicity assay was preformed for multiple commonly used additives. The results of the toxicity assay demonstrate that while salt additives and pH modifications do not significantly affect TX-TL efficiency, solution viscosity and detergent are large factors. In specific, the common cryoprotectant glycerol is sub-optimal. Based on this information, new protein buffers were designed with optimal salt content and DMSO as a cryoprotectant in place of glycerol. | |||
[[Image:toxassay_32813.png|800px]] | |||
Figure 1: Effects of different additives on TX-TL efficiency. A variety of different additives commonly used in protein buffers was tested for toxicity. 25% of the reactions containing the above additive was added to 1nM pBEST-Or2-Or1-Pr-UTR1-deGFP-T500 and endpoint fluorescence after 8 hours was determined – percentages are against a control with no additive internal to the experiment. Error bars represent one standard deviation from three independent experiments. | |||
=== Temperature === | |||
{| border=1 width=100% | |||
|- | |||
| | |||
<center> | |||
[[Image:TXTL-constitutive promoter Figure8.png|600px]] | |||
</center> | |||
<br>'''Figure 8: Transient response of a constitutive promoter as a function of temperature.''' | |||
(A) Schematic illustration and a simple model of a constitutive promoter expressing gfp. | |||
(B) Red blue color map presents preliminary data showing the transient response as a function of temperature. Evaporation dominates at 37 °C. Measurements were acquired over approximately 8 hour duration with indiviudal measurements every 3 minutes. | |||
(C) Red dots represent the dependence of temperature of parameter estimates obtained from this data. These estimates are for the constitutive promoter activity β. | |||
|} | |||
{| border=1 width=100% | |||
|- | |||
| | |||
<center> | |||
[[Image:TXTL-negative feedback Figure9.png|600px]] | |||
</center> | |||
<br> '''Figure 9. Transient response of a negative feedback circuit as a function of temperature.''' | |||
(A) Schematic illustration and simple model of a negative transcriptional feedback circuit based on the autoregulatory effect of the protein fusion TetR-GFP. | |||
(B) Red blue color map presents preliminary data showing the transient response as a function of temperature. Evaporation dominates at 37 °C. Measurements were acquired over approximately 8 hour duration with indiviudal measurements every 3 minutes. | |||
(C) and (D) Red dots represent temperature dependence of parameter estimates obtained from this data. These estimates are for promoter constants β and k. | |||
|} | |||
Revision as of 07:50, 20 May 2013
| Home | Protocols | DNA parts | Preliminary Data | Models | More Info |
This page contains some data that we have taken with the TX-TL breadboard.
Plasmid Expression of GFP
Using pBEST-OR2-OR1-Pr-UTR1-deGFP-T500, a plasmid enhanced for GFP expression, the biomolecular breadboard is able to express mass at equal concentrations to comparable bacteriophage in-vitro systems (J. Shin and V. Noireaux, 2010).
Expression of plasmids can be optimized by concentration. Figure 1 (below) shows a typical time response for expression of the control plasmid, along with the data we collected from our plate reader. If you are trying to get TX-TL working, this is probably the first test that you should do.
Figure 1. eGFP expression as a function of plasmid DNA template. Plasmid DNA pBEST-OR2-OR1-Pr-UTR1-eGFP-T500 is varied by concentration. |
Protecting Linear DNA from Exonuclease-Mediated Degradation
Current standards for circuit design utilize plasmids for DNA template, which require time-consuming subcloning steps. However, circuits based on linear DNA require only PCR assembly or gene synthesis, which drastically decreases preparation time. As a purely extract-derived system, our biomolecular breadboard exhibits exonuclease activity which degrades linear DNA. We are developing multiple technologies to protect linear DNA from exonuclease degradation. These include:
- Protecting linear DNA using noncoding segments
- Inhibiting RecBCD exonuclease with gamS
- Adding thiosulfate bonds to 5' ends
Protection Sequences
The simplest way to protect linear DNA is to put additional "junk" DNA around the construct you are testing. Figure 2 shows the expression curves for different lengths of protection sequences. The overall expression with long protection sequences is about 2X less than what you see with plasmid DNA (compare the plots below to the 2nM plasmid DNA expression levels in Figure 1).
GamS
The GamS protein can be used to protect linear DNA. GamS is a shortened version of the lambda phage Gam protein, which inhibits activity of the exonuclease RecBCD (Murphy, 2007). Figures 3 and 4 show time responses for expression of linear DNA using GamS. Using a combination of GamS and protection sequences, it is possible to get approximately 60% of the expression that you get from plasmid DNA.
|
|
Note that you can include GamS in your reactions either by putting in the DNA encoding GamS or by supplying purified GamS into the reaction. We recommend the latter since using DNA to express GamS uses up some of the resources that would otherwise power your circuit, and also takes a while before you express enough GamS to protect linear DNA.
Thiosulfate Bonds
We have also tried using thiosulfate bonds to protect linear DNA. These have little effect.
Protein Degradation
Implementing protein degradation is integral to enabling the function of many dynamical circuits, such as oscillators or feed-forward loops. We have been exploring the overexpression of AAA ATPases ClpXP to selectively target proteins with degradation tags. Initial experiments indicate that ClpXP, when overexpressed, can accelerate degradation of purified eGFP-ssrA (Fig. 1). We will further characterize this AAA ATPase family, as well as try to demonstrate its use through an incoherent feed-forward loop. We have not been able to replicate the results by adding purified ClpXP protein. However, we intend to express ClpXP off of plasmids or to create custom extract with ClpXP already overexpressed to enable rapid protein degradation.
Environmental Context
The TX-TL system can be used to study the operation of circuits in different environmental contexts, including salts, energy levels and temperature.
Magnesium and Potassium concentration
|
|
Toxicity
To optimize TX-TL for the addition of proteins of interest or reagents (such as gamS for linear protection and ClpX for protein degradation), a toxicity assay was preformed for multiple commonly used additives. The results of the toxicity assay demonstrate that while salt additives and pH modifications do not significantly affect TX-TL efficiency, solution viscosity and detergent are large factors. In specific, the common cryoprotectant glycerol is sub-optimal. Based on this information, new protein buffers were designed with optimal salt content and DMSO as a cryoprotectant in place of glycerol.
Figure 1: Effects of different additives on TX-TL efficiency. A variety of different additives commonly used in protein buffers was tested for toxicity. 25% of the reactions containing the above additive was added to 1nM pBEST-Or2-Or1-Pr-UTR1-deGFP-T500 and endpoint fluorescence after 8 hours was determined – percentages are against a control with no additive internal to the experiment. Error bars represent one standard deviation from three independent experiments.
Temperature
|
|
|
|