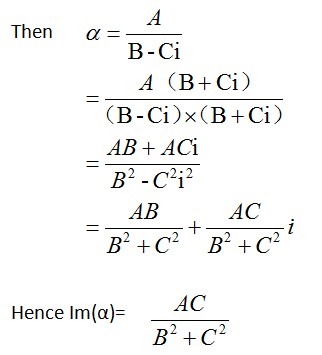Biomod/2014/wiki-2'.html: Difference between revisions
No edit summary |
No edit summary |
||
| (82 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
<html> | <html> | ||
<head> | <head> | ||
<meta http-equiv="Content-Type" content="text/html; charset=utf-8" /> | |||
<style type="text/css"> | |||
body { | |||
display:block; | |||
background-color:#ffffff; | |||
} | |||
} | |||
.ys01 { | |||
text-align: left; | |||
box-shadow: 0px 5px 8px rgba(0,0,0,0.4); | |||
} | |||
} | |||
/* --the header css -- */ | |||
#apDiv1 { | #apDiv1 { | ||
position: | position:relative; | ||
left: | float:left; | ||
top: | left:0px; | ||
width: | top:0px; | ||
height: | margin-right:0px; | ||
width:1000px; | |||
height:360px; | |||
z-index:1; | z-index:1; | ||
} | box-shadow: 0px 5px 8px rgba(0,0,0,0.4); | ||
overflow: visible; | |||
text-align: left; | |||
background-image: url(http://openwetware.org/images/3/37/Header_tianjin.png); | |||
} | |||
/* -- navigator css -- */ | |||
#apDiv2 { | #apDiv2 { | ||
position: | position:relative; | ||
left | float:left; | ||
left:0px; | |||
width: | width:1000px; | ||
height: | height:40px; | ||
z-index:2; | z-index:2; | ||
font-family: Georgia, "Times New Roman", Times, serif; | |||
text-align: left; | |||
background-image: url(http://openwetware.org/images/thumb/7/76/Tianjin_index02.jpg/800px-Tianjin_index02.jpg); | |||
} | |||
#apDiv2 table { | |||
font-size: 18px; | |||
text-align: center; | |||
} | |||
.ys01 #apDiv2 table { | |||
text-align: center; | |||
} | |||
.ys01 #apDiv2 table tr td a { | |||
color: #FFF; | |||
} | |||
/* -- navigator css -- */ | |||
#navDiv li { margin: 0; padding: 0;} | |||
#navDiv ul {margin: 0; padding: 0; list-style: none; z-index:99;} | |||
#navDiv a {text-decoration: none;margin: 0; padding: 0;} | |||
#navDiv | |||
{ | |||
/* -- background body of navigator css -- */ | |||
position: relative; | |||
float: left; | |||
height: 40px; | |||
width:1000px; | |||
left: 0px; | |||
background-color: #ffffff; | |||
background-repeat: repeat-x; | |||
box-shadow: 0px 5px 8px rgba(0,0,0,0.4); | |||
margin-bottom:40px; | |||
z-index:60; | |||
font-family: Georgia, "Times New Roman", Times, serif; | |||
text-align: left; | |||
} | } | ||
# | |||
#navDiv > ul > li { | |||
float: left; | |||
margin-left: 30px; | |||
position: relative; | |||
} | } | ||
# | |||
/* -- when mouse not on the navigator css -- */ | |||
#navDiv > ul > li > a { | |||
color: #333333; | |||
font-family: Verdana, 'Lucida Grande'; | |||
font-size: 15px; | |||
line-height: 40px; | |||
padding: 11px 20px; | |||
-webkit-transition: color .15s; | |||
-moz-transition: color .15s; | |||
-o-transition: color .15s; | |||
transition: color .15s; | |||
} | } | ||
# | |||
/* -- when mouse on the navigator css -- */ | |||
#navDiv > ul > li > a:hover {color: #ffffff; background-color: #000000;} | |||
/* -- top and bottom of the sub meun css -- */ | |||
#navDiv > ul > li > ul | |||
{ | |||
opacity: 0; | |||
visibility: hidden; | |||
padding: 16px 0 20px 0; | |||
background-color: rgb(250,250,250); | |||
text-align: left; | |||
position: absolute; | |||
top: 30px; | |||
left: 50%; | |||
margin-left: -90px; | |||
width: 180px; | |||
-webkit-transition: all 0.3s 0.1s; | |||
-moz-transition: all 0.3s 0.1s; | |||
-o-transition: all 0.3s 0.1s; | |||
transition: all 0.3s 0.1s; | |||
-webkit-border-radius: 5px; | |||
-moz-border-radius: 5px; | |||
border-radius: 5px; | |||
-webkit-box-shadow: 0px 1px 3px rgba(0,0,0,0.4); | |||
-moz-box-shadow: 0px 1px 3px rgba(0,0,0,0.4); | |||
box-shadow: 0px 1px 3px rgba(0,0,0,0.4); | |||
} | } | ||
# | |||
#navDiv > ul > li:hover > ul | |||
{ | |||
opacity: 1; | |||
top: 40px; | |||
visibility: visible; | |||
} | } | ||
# | |||
#navDiv > ul > li > ul:before | |||
{ | |||
content: ''; | |||
display: block; | |||
border-color: transparent transparent rgb(250,250,250) transparent; | |||
border-style: solid; | |||
border-width: 10px; | |||
position: absolute; | |||
top: -20px; | |||
left: 50%; | |||
margin-left: -10px; | |||
} | } | ||
# | |||
#navDiv > ul ul > li { position: relative;} | |||
/* -- when mouse not on the submeun css -- */ | |||
#navDiv ul ul a | |||
{ | |||
color: rgb(50,50,50); | |||
font-family: Verdana, 'Lucida Grande'; | |||
font-size: 13px; | |||
background-color: rgb(250,250,250); | |||
padding: 5px 8px 7px 16px; | |||
display: block; | |||
-webkit-transition: background-color .1s; | |||
-moz-transition: background-color .1s; | |||
-o-transition: background-color .1s; | |||
transition: background-color .1s; | |||
} | } | ||
# | |||
#navDiv ul ul ul | |||
{ | |||
visibility: hidden; | |||
opacity: 0; | |||
position: absolute; | |||
top: -16px; | |||
left: 206px; | |||
padding: 16px 0 20px 0; | |||
background-color: rgb(250,250,250); | |||
text-align: left; | |||
width: 160px; | |||
-webkit-transition: all .3s; | |||
-moz-transition: all .3s; | |||
-o-transition: all .3s; | |||
transition: all .3s; | |||
-webkit-border-radius: 5px; | |||
-moz-border-radius: 5px; | |||
border-radius: 5px; | |||
-webkit-box-shadow: 0px 1px 3px rgba(0,0,0,.4); | |||
-moz-box-shadow: 0px 1px 3px rgba(0,0,0,.4); | |||
box-shadow: 0px 1px 3px rgba(0,0,0,.4); | |||
} | } | ||
#navDiv ul ul > li:hover > ul { opacity: 1; left: 196px; visibility: visible;} | |||
# | /* -- when mouse on the navigator css -- */ | ||
#navDiv ul ul a:hover | |||
{ | |||
background-color: #777777; | |||
color: rgb(240,240,240); | |||
} | |||
} | } | ||
/* Back to top css */ | |||
.back-to { | |||
position: fixed; | |||
bottom: 35px; | |||
*bottom: 50px; | |||
_bottom: 35px; | |||
right: -160px; | |||
z-index: 999; | |||
width: 50px; | |||
zoom: 1; | |||
} | |||
* html .back-to { | |||
/* 不能用 _position 这种写法,因为它在IE7+也会执行expression。。。 */ | |||
position: expression(function(ele){ele.runtimeStyle.position='absolute';Expressions.style.position.fixed(ele);}(this)) | |||
} | |||
.back-to { | |||
float: right; | |||
display: block; | |||
width: 50px; | |||
height: 50px; | |||
background: url(http://a.xnimg.cn/imgpro/button/back-home.png) no-repeat 0 0; | |||
outline: 0 none; | |||
text-indent: -9999em; | |||
} | |||
.back-to:hover { | |||
background-position: -50px 0 | |||
} | |||
.back-to .back-top { | |||
float: right; | |||
display: block; | |||
width: 50px; | |||
height: 50px; | |||
background: url(http://a.xnimg.cn/imgpro/button/back-top.png) no-repeat 0 0; | |||
margin-left: 10px; | |||
outline: 0 none; | |||
text-indent: -9999em; | |||
} | |||
.back-to .backtotop { | |||
float: left; | |||
display: block; | |||
width: 50px; | |||
height: 50px; | |||
background: #666 url(http://a.xnimg.cn/imgpro/arrow/btt.png) 8px -57px no-repeat; | |||
margin-bottom: 15px; | |||
outline: 0 none; | |||
text-indent: -9999em; | |||
-moz-border-radius: 4px; | |||
-khtml-border-radius: 4px; | |||
-webkit-border-radius: 4px; | |||
border-radius: 4px; | |||
position: relative; | |||
-moz-box-shadow: 0px 0px 15px #ccc; | |||
-webkit-box-shadow: 0px 0px 15px #ccc; | |||
box-shadow: 0px 0px 15px #ccc; | |||
} | |||
.back-to .backtotop:hover { | |||
background-color: #333; | |||
background-position: 8px 13px; | |||
} | |||
.back-to .backtotop .back-tip { | |||
position: absolute; | |||
visibility: hidden; | |||
top: -31px; | |||
left: -10px; | |||
} | |||
.back-to .backtotop:hover .back-tip { | |||
visibility: visible; | |||
} | |||
.back-to .back-top:hover { | |||
background-position: -50px 0; | |||
} | |||
#apDiv3 table { | |||
text-align: center; | |||
} | |||
.ys01 #apDiv6 table { | |||
text-align: left; | |||
} | |||
.ys02 { | |||
font-size: 36px; | |||
color: #F00; | |||
} | |||
#apDiv10 { | #apDiv10 { | ||
position:relative; | |||
float:left; | |||
display:block; | |||
left:0px; | |||
width:960px; | |||
z-index:3; | |||
box-shadow: 0px 5px 8px rgba(0,0,0,0.4); | |||
text-align:left; | |||
font-family: Georgia, "Times New Roman", Times, serif; | |||
font-size: 18px; | |||
color: #000; | |||
background-color: #ffffff; | |||
padding: 20px; | |||
} | |||
/*hidden section*/ | |||
.firstHeading{display:none;} | |||
#sidebar-main{display:none;} | |||
#p-cactions{display:none;} | |||
#p-personal{display:none;} | |||
# | |||
} | |||
} | |||
# | |||
} | |||
# | |||
} | |||
# | |||
</style> | </style> | ||
</head> | </head> | ||
< | |||
<body class="ys01";> | |||
<!--header--> | |||
<div id="apDiv1"></div> | |||
<!--navigator starts here--> | |||
<div id="navDiv"> | |||
<ul> | |||
<li class='active '><a href="http://openwetware.org/wiki/Biomod/2014/Tianjin"><span>Home</span></a></li> | |||
<li class='has-sub'><a href="http://openwetware.org/wiki/Biomod/2014/wiki-2%27.html"><span>Idea</span></a> | |||
<ul> | |||
<li><a href='http://openwetware.org/wiki/Biomod/2014/wiki-2%27.html#background'><span>Background</span></a></li> | |||
<li><a href='http://openwetware.org/wiki/Biomod/2014/wiki-2%27.html#motivation'><span>Motivation</span></a></li> | |||
<li><a href='http://openwetware.org/wiki/Biomod/2014/wiki-2%27.html#calculation'><span>Calculation</span></a></li> | |||
</ul> | |||
</li> | |||
<li class='has-sub'><a href="http://openwetware.org/wiki/Biomod/2014/experiment.html"><span>Project</span></a> | |||
<ul> | |||
<li><a href="http://openwetware.org/wiki/Biomod/2014/experiment.html#Three-arm locker"><span>Three-arm Locker</span></a></li> | |||
<li><a href="http://openwetware.org/wiki/Biomod/2014/experiment.html#Gold nanoparticle based photosensor"><span> Gold Nanoparticle Based Photosensor</span></a></li> | |||
<li><a href="http://openwetware.org/wiki/Biomod/2014/experiment.html#dna origami"><span>DNA Origami</span></a></li> | |||
</ul> | |||
</li> | |||
<li class='active'><a href="http://openwetware.org/wiki/Tianjin_protocol"><span>Protocol</span></a> | |||
</li> | |||
<li class='active'><a href="http://openwetware.org/wiki/Biomod/2014/future.html"><span>Future Work</span></a> | |||
</li> | |||
<li class='active '><a href="http://openwetware.org/wiki/Biomod/2014/members.html"><span>Members and Acknowledgement</span></a> | |||
</li> | |||
</ul> | |||
</div> | </div> | ||
<div id=" | |||
<div id="toolBackTo" class="back-to" style="left: 1175px; "> | |||
<a stats="site_footer_back_to_top" class="backtotop" href="#top" onclick="if(Sizzle('#sidebar2 .ready-to-fix')[0]) Sizzle('#sidebar2 .ready-to-fix')[0].style.position = 'static';window.scrollTo(0,0);if(Sizzle('#sidebar2 .ready-to-fix')[0]) Sizzle('#sidebar2 .ready-to-fix')[0].style.position = '';return false;">Back to top | |||
<img stats="site_footer_back_to_top" src="http://openwetware.org/images/a/a7/TJU2012-Back-tip.png" class="back-tip"> | |||
</a> | |||
</div> | </div> | ||
<div id=" | |||
<div id="apDiv10"> | |||
<a name="background"></html> | |||
==Background== | |||
<html></a></html> | |||
<big>DNA Origami</big> | |||
The unique structural motifs and molecular recognition properties of DNA make it a promising template for building nanostructures. Using a long single-stranded DNA as a template, a novel strategy, the so-called DNA origami method, has been developed for the preparation of various two-dimensional (2D) and three-dimensional (3D) nanostructures with defined size which is predictable, precise, controllable and efficient.<br><br> | |||
[[Image:fig02.gif|thumb|550px|center|Fig 1.DNA origami and its applications.<br>]] | |||
DNA origami is an excellent plantform for the nanopatterning and a variety of functional biomolecules and nanoparticles can be assembled onto the DNA origami nanoscaffolds, to obtain complicate nanodevices with special functions which can be used to facilitate imaging, targeted delivery, and controlled release of therapeutic compounds. As described, DNA origami possess the capability of transporting molecular payloads to cells, sensing cell surface inputs for conditional, triggered activation, and reconfiguring its structure for payload delivery. DNA origami structures can also be used as molecular pegboards with a resolution of 4–6 nm, and they been widely used in the assembly of hetero-elements such as proteins and nanoparticles. Therefore, DNA origami has shown great potential in nanotechnology.<br> | |||
<big>Advantages of light</big> | |||
A revolution in cancer therapy has taken place by the emerging use of laser light to achieve controlled and confined thermal damage in the tumor tissue. Laser, the acronym for light amplification by the stimulated emission of radiation, is an optical source that emits photons in a coherent and narrow beam. Laser light has the characteristics of monochromaticity, coherence, and collimation. These properties provide a narrow beam of high intensity, which transmits deep down into the target location with minimal power loss and great precision. <br> | |||
<big>Properties of gold nanoparticles (GNPs)</big> | |||
Nanomedicine is currently an active field because new properties emerge when the size of a matter is reduced from bulk to the nanometer scale. These new properties, including optical, magnetic, electronic, and structural properties, make nano-sized particles (generally 1–100 nm) very promising for a wide range of biomedical applications such as targeted therapy. Plasmonic (noble metal) nanoparticles distinguish themselves from other nanoplatforms by their unique surface plasmon resonance (SPR). A special property of these plasmonic nanoparticles is their heat generation resulting from optical stimulation. <br><br> | |||
[[Image:fig01.gif|thumb|550px|center|Fig 2.The photothermal property for gold nanoparticle.<br>]] | |||
Among plasmonic nanoparticles, gold nanoparticles (GNPs) are most extensively investigated because of their inertness, low cytotoxicity, ready multi-functionalization and long history of medical use. GNPs are also attractive due to their facile synthesis, excellent biocompatibility as well as strongly enhanced and tunable optical properties to convert light into local heat. Gold nanoparticles exhibit light activated photothermal activity due to their geometry dependent SPR. This SPR, resulting from photon confinement to a small particle size, enhances all the radiative and nonradiative properties of GNPs. Hence GNPs have immense potential for the selective laser photothermal therapy of cancer due to their ability to efficiently convert surface plasmon resonance-enhanced absorbed light into localized heat and thus offering multiple modalities for biological and medical applications. <br> | |||
[[Image:那张图.jpg|thumb|550px|center|Fig 3.The the therapy use of gold nanoparticle and DNA origami.<br>]] | |||
<html><a name="motivation"></html> | |||
==Motivation== | |||
<html></a></html> | |||
DNA origami is also emerging as a strong candidate for drug delivery in living systems. For its high compatibility to so many kinds of molecules, such as functional biomolecules and nanoparticles which can be linked to the DNA origami scaffolds, to obtain complicated functional nano-devices. Besides, it's capability to fold structures of 2-dimentional or 3-dimentional makes it will get more circumstances to apply. What’s more, the high rate of development and the endless potential of DNA origami make it worth researching. | |||
But the methods of opening an origami box or changing the structure of it is limited in the addition of ssDNA to open by strand replacement or chemical methods to break, which limits the applications of origami especially in vivo. So, a physical way without adding anything to achieve the goal is what we need and search for. We choose light. But why? | |||
We want to use light to open DNA origami because light control has many advantages. | |||
1. Light is low expenditure and easy to get. | |||
2. It is convenient to control the intensity of light. | |||
3. Light for therapeutic could cause less damage on tissue. | |||
In fact, in treatment, only light will produce sufficient power to penetrate the specific site with less energy loss. And, light has already been used into medical treatment field for many years, such as photothermal therapy. Then, the next question is how to change the light into heat, and the traditional way of photothermal therapy may answer this question. | |||
A type of nanomaterial with strong light absorption comes into the sight of us, which is named gold nanoparticle (Au NP). Au NP has immense potential for the selective laser photothermal therapy of cancer cells due to its photo-thermal activity. It can convert light into heat, and leads to a temperature increase, which serves as our opening trigger. | |||
And as all we know, origami is locked by complementary DNA sequence, which is difficult to totally separate by heating and restoring to room temperature. So that means our origami box have a large possibility to reclose after its open. To overcome this problem, we design three-arm DNA lock to “intelligently” control the open and close of origami by displacement, and it will be showed in the next part. | |||
All in all, by creating and utilizing these principle or design, we want to grant this device with the following control characteristics: | |||
Local release of drugs. Traditional drugs are transported to every corner in human body, and may attack all the cells including the normal ones,such as the low specificity of anticancer drugs. However, our device is supposed to release at a specific location in human body under control. | |||
<big>Selective release of drugs</big> | |||
We want to give our device a DNA logical gate, which enable the device to selectively release specific drugs aimed at curing different cells under different conditions. | |||
<big>Physical control</big> | |||
We replace traditional control methods of adding chemicals with a physical trigger to make our device tunable for future modification, which has never done before. | |||
<html><a name="calculation"></html> | |||
==Calculation== | |||
<html></a></html> | |||
<big>Equation of △T</big> | |||
WHEN | |||
1、the NPs are far enough( interdistance is at least 4 or 5 times larger than the NP diameter ) | |||
2、the NPs are in a homogeneous medium | |||
The temperature increase on the surface of a spherical gold NP can be calculated by using the following equation. | |||
[[Image:2014-08-18_201722.jpg]] | |||
<big>Corrected equation of △T</big> | |||
The fact is that the particle is located close to the interface between two homogeneous media. They are lying on a glass substrate and immersed in water. So the equation has to be corrected by a correction factor b. And κ should be replaced by κeff = κ/b | |||
[[Image:2014-08-18_201723.jpg]] | |||
r is the distance from the center of a particle. We want to calculate the temperature increase on the surface. So r= R, radius of a NP. | |||
[[Image:2014-08-18_201908.jpg]] | |||
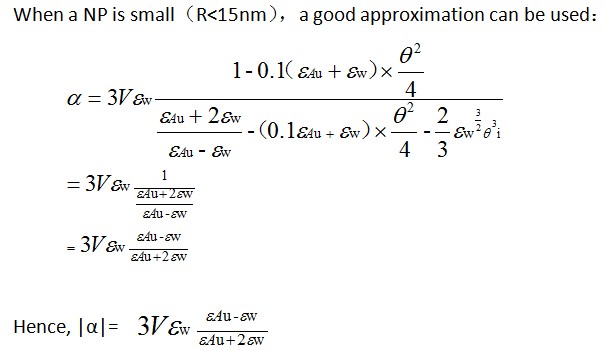
<big>Calculation of σabs</big> | |||
1、when R≤15nm | |||
< | |||
[[Image:2014-08-18_201931.jpg]] | |||
[[Image:2014-08-18_201951.jpg]] | |||
[[Image:2014-08-18_202005.jpg]] | |||
[[Image:2014-08-18_202017.jpg]] | |||
[[Image:2014-08-18_202033.jpg]] | |||
2、when R>15nm | |||
The previous formalism becomes inappropriate and the Mie theory has to be used. Within this model, the σabs is equal to | |||
[[Image:2014-08-18_202737.jpg]] | |||
[[Image:2014-08-18_202813.jpg]] | |||
[[Image:2014-08-18_202827.jpg]] | |||
According to the formulas got above, the dependence amongst the intensity, wave length of the illumination, the size of the nanoparticle and the local temperature increase around a nanoparticle become quantative. Then we are able to select the light source needed for certain temperature increase. For instance, if we use a laser whose wave length is 520nm and intensity is 2.7×10<sup>5</sup>W/cm<sup>-2</sup> to irradiate a 40nm-gold nanoparticle, the local temperature of the nanoparticle will be 70°C. | |||
==References== | |||
[1] Rajendran A, Endo M, Katsuda Y, et al. Programmed two-dimensional self-assembly of multiple DNA origami jigsaw pieces[J]. ACS nano, 2010, 5(1): 665-671.<br><br> | |||
[2] Huang X, Jain P K, El-Sayed I H, et al. Plasmonic photothermal therapy (PPTT) using gold nanoparticles[J]. Lasers in medical science, 2008, 23(3): 217-228.<br><br> | |||
[3] Weissleder R. A clearer vision for in vivo imaging[J]. Nature biotechnology, 2001, 19(4): 316-316. <br><br> | |||
[4] Pinheiro A V, Han D, Shih W M, et al. Challenges and opportunities for structural DNA nanotechnology[J]. Nature nanotechnology, 2011, 6(12): 763-772.<br><br> | |||
[5] Baffou G, Berto P, Bermúdez Ureña E, et al. Photoinduced Heating of Nanoparticle Arrays[J]. ACS nano, 2013, 7(8): 6478-6488.<br><br> | |||
<html> | |||
</div> | </div> | ||
</body> | </body> | ||
</html> | </html> | ||
Latest revision as of 20:46, 25 October 2014
<html> <head> <meta http-equiv="Content-Type" content="text/html; charset=utf-8" /> <style type="text/css">
body {
display:block;
background-color:#ffffff;
}
.ys01 {
text-align: left;
box-shadow: 0px 5px 8px rgba(0,0,0,0.4);
}
/* --the header css -- */
- apDiv1 {
position:relative;
float:left;
left:0px; top:0px;
margin-right:0px;
width:1000px; height:360px; z-index:1;
box-shadow: 0px 5px 8px rgba(0,0,0,0.4);
overflow: visible; text-align: left; background-image: url(http://openwetware.org/images/3/37/Header_tianjin.png);
}
/* -- navigator css -- */
- apDiv2 {
position:relative; float:left;
left:0px;
width:1000px; height:40px; z-index:2; font-family: Georgia, "Times New Roman", Times, serif; text-align: left; background-image: url(http://openwetware.org/images/thumb/7/76/Tianjin_index02.jpg/800px-Tianjin_index02.jpg);
}
- apDiv2 table {
font-size: 18px;
text-align: center;
}
.ys01 #apDiv2 table {
text-align: center;
}
.ys01 #apDiv2 table tr td a { color: #FFF;
}
/* -- navigator css -- */
- navDiv li { margin: 0; padding: 0;}
- navDiv ul {margin: 0; padding: 0; list-style: none; z-index:99;}
- navDiv a {text-decoration: none;margin: 0; padding: 0;}
- navDiv
{
/* -- background body of navigator css -- */
position: relative;
float: left;
height: 40px; width:1000px; left: 0px; background-color: #ffffff; background-repeat: repeat-x; box-shadow: 0px 5px 8px rgba(0,0,0,0.4); margin-bottom:40px;
z-index:60;
font-family: Georgia, "Times New Roman", Times, serif;
text-align: left;
}
- navDiv > ul > li {
float: left; margin-left: 30px; position: relative;
}
/* -- when mouse not on the navigator css -- */
- navDiv > ul > li > a {
color: #333333; font-family: Verdana, 'Lucida Grande'; font-size: 15px; line-height: 40px; padding: 11px 20px;
-webkit-transition: color .15s;
-moz-transition: color .15s;
-o-transition: color .15s;
transition: color .15s;
}
/* -- when mouse on the navigator css -- */
- navDiv > ul > li > a:hover {color: #ffffff; background-color: #000000;}
/* -- top and bottom of the sub meun css -- */
- navDiv > ul > li > ul
{
opacity: 0; visibility: hidden; padding: 16px 0 20px 0; background-color: rgb(250,250,250); text-align: left; position: absolute; top: 30px; left: 50%; margin-left: -90px; width: 180px;
-webkit-transition: all 0.3s 0.1s;
-moz-transition: all 0.3s 0.1s;
-o-transition: all 0.3s 0.1s;
transition: all 0.3s 0.1s;
-webkit-border-radius: 5px;
-moz-border-radius: 5px;
border-radius: 5px;
-webkit-box-shadow: 0px 1px 3px rgba(0,0,0,0.4);
-moz-box-shadow: 0px 1px 3px rgba(0,0,0,0.4);
box-shadow: 0px 1px 3px rgba(0,0,0,0.4);
}
- navDiv > ul > li:hover > ul
{
opacity: 1; top: 40px; visibility: visible;
}
- navDiv > ul > li > ul:before
{
content: ''; display: block; border-color: transparent transparent rgb(250,250,250) transparent; border-style: solid; border-width: 10px; position: absolute; top: -20px; left: 50%; margin-left: -10px;
}
- navDiv > ul ul > li { position: relative;}
/* -- when mouse not on the submeun css -- */
- navDiv ul ul a
{
color: rgb(50,50,50); font-family: Verdana, 'Lucida Grande'; font-size: 13px; background-color: rgb(250,250,250); padding: 5px 8px 7px 16px; display: block;
-webkit-transition: background-color .1s;
-moz-transition: background-color .1s;
-o-transition: background-color .1s;
transition: background-color .1s;
}
- navDiv ul ul ul
{
visibility: hidden; opacity: 0; position: absolute; top: -16px; left: 206px; padding: 16px 0 20px 0; background-color: rgb(250,250,250); text-align: left; width: 160px;
-webkit-transition: all .3s;
-moz-transition: all .3s;
-o-transition: all .3s;
transition: all .3s;
-webkit-border-radius: 5px;
-moz-border-radius: 5px;
border-radius: 5px;
-webkit-box-shadow: 0px 1px 3px rgba(0,0,0,.4);
-moz-box-shadow: 0px 1px 3px rgba(0,0,0,.4);
box-shadow: 0px 1px 3px rgba(0,0,0,.4);
}
- navDiv ul ul > li:hover > ul { opacity: 1; left: 196px; visibility: visible;}
/* -- when mouse on the navigator css -- */
- navDiv ul ul a:hover
{
background-color: #777777; color: rgb(240,240,240);
}
/* Back to top css */
.back-to {
position: fixed; bottom: 35px; *bottom: 50px; _bottom: 35px; right: -160px; z-index: 999; width: 50px; zoom: 1; }
- html .back-to {
/* 不能用 _position 这种写法,因为它在IE7+也会执行expression。。。 */
position: expression(function(ele){ele.runtimeStyle.position='absolute';Expressions.style.position.fixed(ele);}(this))
}
.back-to {
float: right; display: block; width: 50px; height: 50px; background: url(http://a.xnimg.cn/imgpro/button/back-home.png) no-repeat 0 0; outline: 0 none; text-indent: -9999em;
}
.back-to:hover {
background-position: -50px 0 }
.back-to .back-top {
float: right; display: block; width: 50px; height: 50px; background: url(http://a.xnimg.cn/imgpro/button/back-top.png) no-repeat 0 0; margin-left: 10px; outline: 0 none; text-indent: -9999em; }
.back-to .backtotop {
float: left; display: block; width: 50px; height: 50px; background: #666 url(http://a.xnimg.cn/imgpro/arrow/btt.png) 8px -57px no-repeat; margin-bottom: 15px; outline: 0 none; text-indent: -9999em; -moz-border-radius: 4px; -khtml-border-radius: 4px; -webkit-border-radius: 4px; border-radius: 4px; position: relative; -moz-box-shadow: 0px 0px 15px #ccc; -webkit-box-shadow: 0px 0px 15px #ccc; box-shadow: 0px 0px 15px #ccc; }
.back-to .backtotop:hover {
background-color: #333; background-position: 8px 13px; }
.back-to .backtotop .back-tip {
position: absolute; visibility: hidden; top: -31px; left: -10px; }
.back-to .backtotop:hover .back-tip {
visibility: visible; }
.back-to .back-top:hover {
background-position: -50px 0; }
- apDiv3 table {
text-align: center;
}
.ys01 #apDiv6 table { text-align: left;
}
.ys02 {
font-size: 36px;
color: #F00;
}
- apDiv10 {
position:relative;
float:left;
display:block;
left:0px;
width:960px; z-index:3;
box-shadow: 0px 5px 8px rgba(0,0,0,0.4);
text-align:left; font-family: Georgia, "Times New Roman", Times, serif; font-size: 18px; color: #000; background-color: #ffffff;
padding: 20px;
}
/*hidden section*/
.firstHeading{display:none;}
- sidebar-main{display:none;}
- p-cactions{display:none;}
- p-personal{display:none;}
</style> </head>
<body class="ys01";>
<!--header--> <div id="apDiv1"></div>
<!--navigator starts here-->
<div id="navDiv">
<ul>
<li class='active '><a href="http://openwetware.org/wiki/Biomod/2014/Tianjin"><span>Home</span></a></li>
<li class='has-sub'><a href="http://openwetware.org/wiki/Biomod/2014/wiki-2%27.html"><span>Idea</span></a>
<ul>
<li><a href='http://openwetware.org/wiki/Biomod/2014/wiki-2%27.html#background'><span>Background</span></a></li>
<li><a href='http://openwetware.org/wiki/Biomod/2014/wiki-2%27.html#motivation'><span>Motivation</span></a></li>
<li><a href='http://openwetware.org/wiki/Biomod/2014/wiki-2%27.html#calculation'><span>Calculation</span></a></li>
</ul>
</li>
<li class='has-sub'><a href="http://openwetware.org/wiki/Biomod/2014/experiment.html"><span>Project</span></a>
<ul>
<li><a href="http://openwetware.org/wiki/Biomod/2014/experiment.html#Three-arm locker"><span>Three-arm Locker</span></a></li>
<li><a href="http://openwetware.org/wiki/Biomod/2014/experiment.html#Gold nanoparticle based photosensor"><span> Gold Nanoparticle Based Photosensor</span></a></li>
<li><a href="http://openwetware.org/wiki/Biomod/2014/experiment.html#dna origami"><span>DNA Origami</span></a></li>
</ul>
</li>
<li class='active'><a href="http://openwetware.org/wiki/Tianjin_protocol"><span>Protocol</span></a>
</li>
<li class='active'><a href="http://openwetware.org/wiki/Biomod/2014/future.html"><span>Future Work</span></a>
</li>
<li class='active '><a href="http://openwetware.org/wiki/Biomod/2014/members.html"><span>Members and Acknowledgement</span></a>
</li>
</ul> </div>
<div id="toolBackTo" class="back-to" style="left: 1175px; ">
<a stats="site_footer_back_to_top" class="backtotop" href="#top" onclick="if(Sizzle('#sidebar2 .ready-to-fix')[0]) Sizzle('#sidebar2 .ready-to-fix')[0].style.position = 'static';window.scrollTo(0,0);if(Sizzle('#sidebar2 .ready-to-fix')[0]) Sizzle('#sidebar2 .ready-to-fix')[0].style.position = '';return false;">Back to top
<img stats="site_footer_back_to_top" src="http://openwetware.org/images/a/a7/TJU2012-Back-tip.png" class="back-tip">
</a>
</div>
<div id="apDiv10"> <a name="background"></html>
Background
<html></a></html> DNA Origami
The unique structural motifs and molecular recognition properties of DNA make it a promising template for building nanostructures. Using a long single-stranded DNA as a template, a novel strategy, the so-called DNA origami method, has been developed for the preparation of various two-dimensional (2D) and three-dimensional (3D) nanostructures with defined size which is predictable, precise, controllable and efficient.

DNA origami is an excellent plantform for the nanopatterning and a variety of functional biomolecules and nanoparticles can be assembled onto the DNA origami nanoscaffolds, to obtain complicate nanodevices with special functions which can be used to facilitate imaging, targeted delivery, and controlled release of therapeutic compounds. As described, DNA origami possess the capability of transporting molecular payloads to cells, sensing cell surface inputs for conditional, triggered activation, and reconfiguring its structure for payload delivery. DNA origami structures can also be used as molecular pegboards with a resolution of 4–6 nm, and they been widely used in the assembly of hetero-elements such as proteins and nanoparticles. Therefore, DNA origami has shown great potential in nanotechnology.
Advantages of light
A revolution in cancer therapy has taken place by the emerging use of laser light to achieve controlled and confined thermal damage in the tumor tissue. Laser, the acronym for light amplification by the stimulated emission of radiation, is an optical source that emits photons in a coherent and narrow beam. Laser light has the characteristics of monochromaticity, coherence, and collimation. These properties provide a narrow beam of high intensity, which transmits deep down into the target location with minimal power loss and great precision.
Properties of gold nanoparticles (GNPs)
Nanomedicine is currently an active field because new properties emerge when the size of a matter is reduced from bulk to the nanometer scale. These new properties, including optical, magnetic, electronic, and structural properties, make nano-sized particles (generally 1–100 nm) very promising for a wide range of biomedical applications such as targeted therapy. Plasmonic (noble metal) nanoparticles distinguish themselves from other nanoplatforms by their unique surface plasmon resonance (SPR). A special property of these plasmonic nanoparticles is their heat generation resulting from optical stimulation.

Among plasmonic nanoparticles, gold nanoparticles (GNPs) are most extensively investigated because of their inertness, low cytotoxicity, ready multi-functionalization and long history of medical use. GNPs are also attractive due to their facile synthesis, excellent biocompatibility as well as strongly enhanced and tunable optical properties to convert light into local heat. Gold nanoparticles exhibit light activated photothermal activity due to their geometry dependent SPR. This SPR, resulting from photon confinement to a small particle size, enhances all the radiative and nonradiative properties of GNPs. Hence GNPs have immense potential for the selective laser photothermal therapy of cancer due to their ability to efficiently convert surface plasmon resonance-enhanced absorbed light into localized heat and thus offering multiple modalities for biological and medical applications.

<html><a name="motivation"></html>
Motivation
<html></a></html>
DNA origami is also emerging as a strong candidate for drug delivery in living systems. For its high compatibility to so many kinds of molecules, such as functional biomolecules and nanoparticles which can be linked to the DNA origami scaffolds, to obtain complicated functional nano-devices. Besides, it's capability to fold structures of 2-dimentional or 3-dimentional makes it will get more circumstances to apply. What’s more, the high rate of development and the endless potential of DNA origami make it worth researching.
But the methods of opening an origami box or changing the structure of it is limited in the addition of ssDNA to open by strand replacement or chemical methods to break, which limits the applications of origami especially in vivo. So, a physical way without adding anything to achieve the goal is what we need and search for. We choose light. But why?
We want to use light to open DNA origami because light control has many advantages.
1. Light is low expenditure and easy to get.
2. It is convenient to control the intensity of light.
3. Light for therapeutic could cause less damage on tissue.
In fact, in treatment, only light will produce sufficient power to penetrate the specific site with less energy loss. And, light has already been used into medical treatment field for many years, such as photothermal therapy. Then, the next question is how to change the light into heat, and the traditional way of photothermal therapy may answer this question.
A type of nanomaterial with strong light absorption comes into the sight of us, which is named gold nanoparticle (Au NP). Au NP has immense potential for the selective laser photothermal therapy of cancer cells due to its photo-thermal activity. It can convert light into heat, and leads to a temperature increase, which serves as our opening trigger.
And as all we know, origami is locked by complementary DNA sequence, which is difficult to totally separate by heating and restoring to room temperature. So that means our origami box have a large possibility to reclose after its open. To overcome this problem, we design three-arm DNA lock to “intelligently” control the open and close of origami by displacement, and it will be showed in the next part.
All in all, by creating and utilizing these principle or design, we want to grant this device with the following control characteristics: Local release of drugs. Traditional drugs are transported to every corner in human body, and may attack all the cells including the normal ones,such as the low specificity of anticancer drugs. However, our device is supposed to release at a specific location in human body under control.
Selective release of drugs
We want to give our device a DNA logical gate, which enable the device to selectively release specific drugs aimed at curing different cells under different conditions.
Physical control
We replace traditional control methods of adding chemicals with a physical trigger to make our device tunable for future modification, which has never done before.
<html><a name="calculation"></html>
Calculation
<html></a></html> Equation of △T
WHEN
1、the NPs are far enough( interdistance is at least 4 or 5 times larger than the NP diameter )
2、the NPs are in a homogeneous medium
The temperature increase on the surface of a spherical gold NP can be calculated by using the following equation.
Corrected equation of △T
The fact is that the particle is located close to the interface between two homogeneous media. They are lying on a glass substrate and immersed in water. So the equation has to be corrected by a correction factor b. And κ should be replaced by κeff = κ/b
r is the distance from the center of a particle. We want to calculate the temperature increase on the surface. So r= R, radius of a NP.
Calculation of σabs
1、when R≤15nm
2、when R>15nm
The previous formalism becomes inappropriate and the Mie theory has to be used. Within this model, the σabs is equal to
According to the formulas got above, the dependence amongst the intensity, wave length of the illumination, the size of the nanoparticle and the local temperature increase around a nanoparticle become quantative. Then we are able to select the light source needed for certain temperature increase. For instance, if we use a laser whose wave length is 520nm and intensity is 2.7×105W/cm-2 to irradiate a 40nm-gold nanoparticle, the local temperature of the nanoparticle will be 70°C.
References
[1] Rajendran A, Endo M, Katsuda Y, et al. Programmed two-dimensional self-assembly of multiple DNA origami jigsaw pieces[J]. ACS nano, 2010, 5(1): 665-671.
[2] Huang X, Jain P K, El-Sayed I H, et al. Plasmonic photothermal therapy (PPTT) using gold nanoparticles[J]. Lasers in medical science, 2008, 23(3): 217-228.
[3] Weissleder R. A clearer vision for in vivo imaging[J]. Nature biotechnology, 2001, 19(4): 316-316.
[4] Pinheiro A V, Han D, Shih W M, et al. Challenges and opportunities for structural DNA nanotechnology[J]. Nature nanotechnology, 2011, 6(12): 763-772.
[5] Baffou G, Berto P, Bermúdez Ureña E, et al. Photoinduced Heating of Nanoparticle Arrays[J]. ACS nano, 2013, 7(8): 6478-6488.
<html>
</div>
</body>
</html>