Biomod/2014/UCR/Breaking RNA/Acknowledgements
<html lang=""> <head>
<meta charset='utf-8'> <meta http-equiv="X-UA-Compatible" content="IE=edge"> <meta name="viewport" content="width=device-width, initial-scale=1"> <link rel="stylesheet" href="styles.css"> <script src="http://code.jquery.com/jquery-latest.min.js" type="text/javascript"></script> <script src="script.js"></script> <title>Breaking RNA</title>
<style>
- column-one,.toc {
display: none;
} body.mediawiki {
background: url(http://openwetware.org/images/8/8c/Breaking_RNA_Background.png); background-size: cover;
}
- globalWrapper{
width: 80%; margin-left: 10%; position: absolute;
}
- content{
margin: 0px;
<!-- background: #e6e6e6; -->
background: white;
}
h1,h2,h3,p,ul,li {
font-family: 'Open Sans', sans-serif; font-weight: 300; color: Black; text-align: justify;
} h1
{
text-align: center;
}
h3
{
font-size: 20px;
position: relative;
text-align:center;
}
p
{
font-size: 16px;
line-height: 30px;
padding: 10px 150px;
}
- objectives
{
}
- objectives ul
{
padding: 0 0 0 30%;
margin: 0 auto;
}
- objectives li
{
font-size: 18px;
list-style: none;
line-height: 30px;
}
- submenu
{
margin: 0px 40px 0px 22px;
}
- submenu a
{
color: black;
text-decoration: underline;
}
- submenu a:hover
{
color: rgba(255,255,255,0.99);
}
- submenu ul
{
text-align: center;
padding: 8px 0px;
border: 3px solid black;
background: rgba(70,150,70,0.8);
<!--background: url(http://openwetware.org/images/8/8c/Breaking_RNA_Background.png);
background-size: cover;-->
}
- submenu li
{
font-size: 18px;
color: black;
padding: 0px 20px;
display: inline-block;
}
- supplemenu
{
margin: 0px 40px 0px 22px;
}
- supplemenu a
{
color: black;
text-decoration: underline;
}
- supplemenu a:hover
{
color: rgba(255,255,255,0.99);
}
- supplemenu ul
{
text-align: center;
padding: 15px 0px;
border: 3px solid black;
background: rgba(70,150,70,0.8);
}
- supplemenu li
{
font-size: 18px;
color: black;
padding: 0px 10px;
display: inline-block;
}
- floatmenu
{
position: fixed;
bottom: 0;
width: 78%;
font-size: 18px;
color: black;
padding: 0px 10px;
background: rgba(70,150,70,1);
border: 3px solid black;
}
- floatmenu td
{
text-align: center;
height: 36px;
}
- floatmenu td:hover
{
background: white;
}
- button
{
border-left: 3px solid black;
}
- floatmenu a
{
text-decoration: none;
color: black;
}
@import url(http://fonts.googleapis.com/css?family=Open+Sans:700);
#cssmenu {
background: url(http://openwetware.org/images/8/8c/Breaking_RNA_Background.png); background-size: cover; width: auto; border: 3px solid rgba(0,0,0,0.8);
} #cssmenu ul { list-style: none; margin: 0; padding: 0; line-height: 1;
text-align:center;
display: block; zoom: 1; } #cssmenu ul:after { content: " "; display: block; font-size: 0; height: 0; clear: both; visibility: hidden; } #cssmenu ul li { display: inline-block; padding: 0; margin: 0;
line-height: 1;
} #cssmenu.align-right ul li { float: right; } #cssmenu.align-center ul { text-align: center; } #cssmenu ul li a { color: #ffffff; text-decoration: none; display: block; padding: 15px 25px; font-family: 'Open Sans', sans-serif; font-weight: 300; text-transform: uppercase; font-size: 14px; position: relative; -webkit-transition: color .25s; -moz-transition: color .25s; -ms-transition: color .25s; -o-transition: color .25s; transition: color .25s; } #cssmenu ul li a:hover { color: #f2ff00; } #cssmenu ul li a:hover:before { width: 100%; } #cssmenu ul li a:after { content: ""; display: block; position: absolute; right: -3px; top: 19px; height: 6px; width: 6px; background: #ffffff; opacity: .5; } #cssmenu ul li a:before { content: ""; display: block; position: absolute; left: 0; bottom: 0; height: 3px; width: 0; background: #f2ff00; -webkit-transition: width .25s; -moz-transition: width .25s; -ms-transition: width .25s; -o-transition: width .25s; transition: width .25s; } #cssmenu ul li.last > a:after, #cssmenu ul li:last-child > a:after { display: none; } #cssmenu ul li.active a { color: #f2ff00; } #cssmenu ul li.active a:before { width: 100%; } #cssmenu.align-right li.last > a:after, #cssmenu.align-right li:last-child > a:after { display: block; } #cssmenu.align-right li:first-child a:after { display: none; } @media screen and (max-width: 768px) {
#supplemenu ul li {
float: none; display: block; } #supplemenu ul li a { width: 100%; } #cssmenu ul li { float: none; display: block; } #cssmenu ul li a { width: 100%; -moz-box-sizing: border-box; -webkit-box-sizing: border-box; box-sizing: border-box; border-bottom: 1px solid #ffffff; } #cssmenu ul li.last > a, #cssmenu ul li:last-child > a { border: 0; } #cssmenu ul li a:after { display: none; } #cssmenu ul li a:before { display: none; } }
</style>
</head>
<body>
</body>
</html>
Supplement
Oligonucleotide Sequences
1.1 Oscillator Genes and Strands
DNA Oligonucleotides Base Pair Length Sequences G1 Non-Template 83 5'-TAA TAC GAC TCA CTA TAG GAT GGC AGC GGA GAG TTG CTT GGA ATG CGT TAT AGT CTC TTA GGT GTG TTC GCA CAC CAC TCT CC-3' G1 Template 83 5'-GGA GAG TGG TGT GCG AAC ACA CCT AAG AGA CTA TAA CGC ATT CCA AGC AAC TCT CCG CTG CCA TCC TAT AGT GAG TCG TAT TA-3' G2 Non-Template 56 5'-ATT TAG GTG ACA CTA TAG AGG CGA GAA GCA GCA ATG ATA GTG GAA TTG ACT TAC GC-3' G2 Template 56 5'-GCG TAA GTC AAT TCC ACT ATC ATT GCT GCT TCT CGC CTC TAT AGT GTC ACC TAA AT-3' G3 Non-Template 52 5'-TTC TAA TAC GAC TCA CTA TAG CGT AAG TCA ATT CCA CTA TCA TTG CTG CTT C-3' G3 Template 52 5'-GAA GCA GCA ATG ATA GTG GAA TTG ACT TAC GCT ATA GTG AGT CGT ATT AGA A-3' G4 Non-Template 83 5'-ATT TAG GTG ACA CTA TAG AGG CGA CGT CGC CTC TCA ACG AAC CTT ACG CAA TAT CAG AGA ATC CAC ACA AGC GTG TGG TGA GAG G-3' G4 Template 83 5'-CCT CTC ACC ACA CGC TTG TGT GGA TTC TCT GAT ATT GCG TAA GGT TCG TTG AGA GGC GAC GTC GCC TCT ATA GTG TCA CCT AAA T-3' k1 61 5'-TexRd-CGT CGC CTC TCA ACG AAC CTT ACG CAA TAT CAG AGA ATC CAC ACA AGC GTG TGG TGA GAG G-IowaBlack-3'
1.2 Bistable Genes and Strands
DNA Oligonucleotides Base Pair Length Sequences G1 Non-Template 83 5'-TAA TAC GAC TCA CTA TAG GAT GGC AGC GGA GAG TTG CTT GGA ATG CGT TAT AGT CTC TTA GGT GTG TTC GCA CAC CAC TCT CC-3' G1 Template 83 5'-GGA GAG TGG TGT GCG AAC ACA CCT AAG AGA CTA TAA CGC ATT CCA AGC AAC TCT CCG CTG CCA TCC TAT AGT GAG TCG TAT TA-3' G2(B) Non-Template 57 5'-ATT TAG GTG ACA CTA TAG AGG CGA GCG TAA GTC AAT TCC ACT ATC ATT GCT GCA AGC-3' G2(B) Template 57 5'-GCT TGC AGC AAT GAT AGT GGA ATT GAC TTA CGC TCG CCT CTA TAG TGT CAC CTA AAT-3' k1 100 5'-TexRd-CGT CGC CTC TCA ACG AAC CTT ACG CAA TAT CAG AGA ATC CAC ACA AGC GTG TGG TGA GAG G-IowaBlack-3' Spinach Non-Template 100 5'-ATT TAG GTG ACA CTA TAG AGG ACG CGA CCG AAA TGG TGA AGG ACG GGT CCA GTG CTT CGG CAC TGT TGA GTA GAG TGT GAG CTC CGT AAC TGG TCG CGT C-3' Spinach Template 100 5'-GAC GCG ACC AGT TAC GGA GCT CAC ACT CTA CTC AAC AGT GCC GAA GCA CTG GAC CCG TCC TTC ACC ATT TCG GTC GCG TCC TCT ATA GTG TCA CCT AAA T-3' Malachite Green Non-Template 68 5'-ACT ATG ATA ATA CGA CTC ACT ATA GGG AGA GGA TCC CGA CTG GCG AGA GCC AGG TAA CGA ATG GAT CC-3' Malachite Green Template 68 5'-GGA TCC ATT CGT TAC CTG GCT CTC GCC AGT CGG GAT CCT CTC CCT ATA GTG AGT CGT ATT ATC ATA GT-3'
Characterization of Spinach and Malachite Green aptamer
Binding of MG and SP Aptamers


Unbound Aptamer-Kleptamer Interaction
An important factor in the oscillatory system is to ensure the R1 and K1 strands are not strongly interacting before the inhibition of SP6 RNAP. In this gel, the R1 and K1 aptamers were mixed into solution together at the same concentration and incubated. For both variations of the K1 strand (23&38 bp), there are two distinct bands in lanes 4 and 6. This indicates that there are little interactions occurring between these two strands before R1 binds to the enzyme. This can be explained by a difference in secondary structure between the bound and unbound forms of R1.

Reactivation of T7 RNA Polymerase with Genelets
EDIT
In this case, the genes- G3 and G2 –were used instead of the aptamers. There is obvious inhibition of the enzyme with the addition of varying concentrations of G3. After a few hours, G2 (750 nM) was added to the mixture to be transcribed into the reactivator, R2. Unfortunately, there was no reactivation of the enzyme. It’s possible this may be because the transcription of G3 is outcompeting the transcription of G2. This would prevent any reactivation since T7 RNAP would immediately re-inhibit itself.

Comparison of Transcription Rates between SP6 RNAP and T7 RNAP

Modeling
Switch


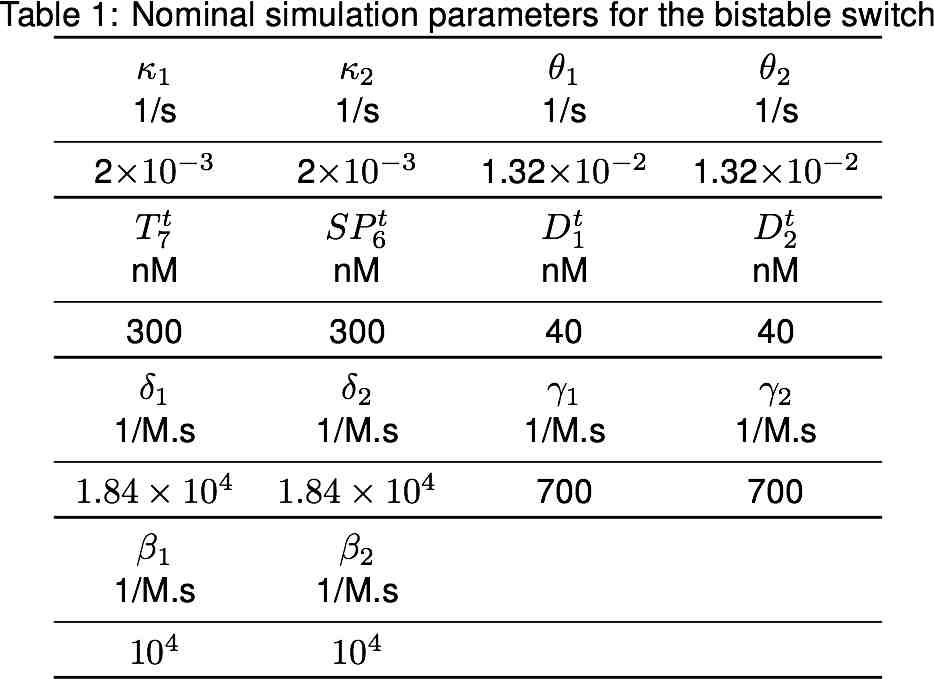
Clock


