Biomod/2013/UT-Austin/Design: Difference between revisions
(→Design) |
(→Design) |
||
| Line 14: | Line 14: | ||
In order to have a circuit that would specifically detect and characterize SNP309 of the MDM2 gene, new CHA circuit pieces were designed using CircDesigNA. Hairpins were designed to not interact with the MDM2 gene in the absence of the split catalyst pieces, '''Table 1'''. The split catalyst was designed as depicted in '''Figure 10'''. The MDM2 gene sequence determined a majority of the split catalyst sequences and the remaining sequences were designed by CircDesigNA. | In order to have a circuit that would specifically detect and characterize SNP309 of the MDM2 gene, new CHA circuit pieces were designed using CircDesigNA. Hairpins were designed to not interact with the MDM2 gene in the absence of the split catalyst pieces, '''Table 1'''. The split catalyst was designed as depicted in '''Figure 10'''. The MDM2 gene sequence determined a majority of the split catalyst sequences and the remaining sequences were designed by CircDesigNA. | ||
[[Image:mdm2_target.png|thumb| | [[Image:mdm2_target.png|thumb|250px|center|'''Figure 10:''' The MDM2 SNP is our detection target and acts as an '''mOS''' for the purposes of scCHA. The catalyst piece containing the 1 domain (red) is complementary to the region of the MDM2 gene upstream of SNP309, p0. This complementarity continues into the first two bases of 1. The catalyst piece with the 2 and 3 domains contain a region that is complementary to the region downstream of SNP309. These complementary regions bring both catalyst pieces together for CHA triggering.]] | ||
{| class="wikitable" border="1" | {| class="wikitable" border="1" | ||
| Line 60: | Line 60: | ||
The apurinic circuit was designed in the same fashion as the MDM2 circuit. The catalyst pieces were designed to assemble on the A4324 loop of the 28S rRNA, '''Figure 11'''. The remaining circuit pieces for the apurinic design can be found in '''Table 1'''. | The apurinic circuit was designed in the same fashion as the MDM2 circuit. The catalyst pieces were designed to assemble on the A4324 loop of the 28S rRNA, '''Figure 11'''. The remaining circuit pieces for the apurinic design can be found in '''Table 1'''. | ||
[[Image:ap_target.png|thumb| | [[Image:ap_target.png|thumb|250px|center|'''Figure 11:''' The catalyst pieces assemble on the A4234 loop of the 28S rRNA such that the A4324 ( p0) is the complement of the first base of domain 1.]] | ||
Revision as of 23:38, 26 October 2013
<html> <style type="text/css"> //<!--
/* Define the background of the whole page */
@media screen {
body { background: #c5d0ea 0 0 no-repeat; }
} /* Define the div holding the page contents */
- globalWrapper{
left: 50%; height: 100%; background-color: #FFFFFF; width: 900px; margin-left: -450px; /* Should be width / 2 */ margin-top: 0px; margin-bottom: 0px; margin-right: 0px; padding: 0 0 0 0; position: absolute;
} .toc{
display:none;
} /* Hide the page heading at top */ .firstHeading {display:none;} /* Hide the bar at the left */
- column-one {display:none;}
/* Define the div holding the page contents (this div is inside globalWrapper. We override its default margins) */
- content{
margin: 0 0 0 0; font: normal 100% sans-serif; color: #000033; padding: 12px 12px 12px 12px; background-image: url()" ; background-repeat: no-repeat; border: 0;
} /* Miscellaneous stuff. Feel free to add style definitions. */ /* h3 {
/* color: #CC5500; font: normal 3em sans-serif; letter-spacing: 1px; margin-bottom: 0; */ color: #000000; font-style: normal; font-size: 39px; font-family: 'Playbill2', 'Playbill', sans-serif; //font-stretch: ultra-expanded; letter-spacing: 1px; margin-bottom: -10px; line-height: 25px; padding-bottom: 0px;
}
- /
h2 {
color: #000000; font-style: normal; font-size: 39px; font-family: 'Montaga', serif; //font-stretch: ultra-expanded; letter-spacing: 1px; margin-bottom: -10px; line-height: 25px; padding-bottom: 0px;
} h1 {
/* color: #CC5500; font: normal 3em Montaga; letter-spacing: 1px; margin-bottom: 0; */ color: #000000; font-style: normal; font-size: 39px; font-family: 'Montaga', serif; //font-stretch: ultra-expanded; letter-spacing: 1px; margin-bottom: -10px; line-height: 25px; padding-bottom: 0px;
}
.scriptFont {
color: #0; padding-top: 5px; font: normal 1em Georgia;
}
//-->
</style>
<div style="text-align:center" />
<a href="http://openwetware.org/wiki/Biomod/2013/UT-Austin"><img src="http://openwetware.org/images/2/2f/G4262.png" width ="50%" height="50%"/><br />
<!-- links at top -->
<div class="linkconcept linkgeneral">
<a href="http://openwetware.org/wiki/Biomod/2013/UT-Austin/Introduction"><img src="http://openwetware.org/images/0/0d/Intro.png" width="20%" alt="" /></a>
</div> <div class="linkdesign linkgeneral">
<a href="http://openwetware.org/wiki/Biomod/2013/UT-Austin/Design"><img src="http://openwetware.org/images/0/0a/Design.png" width="10%" alt="" /></a>
</div> <div class="linkmethods linkgeneral">
<a href="http://openwetware.org/wiki/Biomod/2013/UT-Austin/Methods"><img src="http://openwetware.org/images/0/07/MEthods.png" width="15%" alt="" /></a>
</div> <div class="linkresults linkgeneral">
<a href="http://openwetware.org/wiki/Biomod/2013/UT-Austin/Results"><img src="http://openwetware.org/images/1/17/Results.png" width="12%" alt="" /></a>
</div> <div class="linkmembers linkgeneral">
<a href="http://openwetware.org/wiki/Biomod/2013/UT-Austin/Team Info"><img src="http://openwetware.org/images/3/32/Team.png" width="15%" alt="" /></a>
</div> <div class="linkrefs linkgeneral">
<a href="http://openwetware.org/wiki/Biomod/2013/UT-Austin/References"><img src="http://openwetware.org/images/0/0d/References.png" width="20%" alt="" /></a>
</div> <div style="clear: both;" ></div>
</div>
</html>
Design
The CHA reaction described in the introduction allows for the detection of oligonucleotides. The oligo detected is C1, Figure 8A. C1 has 3 domains: the toehold binding domain (1*) and the domains that undergo branch migration in order to open H1. However, previous work has shown that these catalytic domains do not have to be found on a single oligo. These domains can be split between separate oligos and still trigger CHA when colocalized on a common template, Figure 8B. This modified variant of CHA will be referred to as split catalyst CHA (scCHA) in this work.
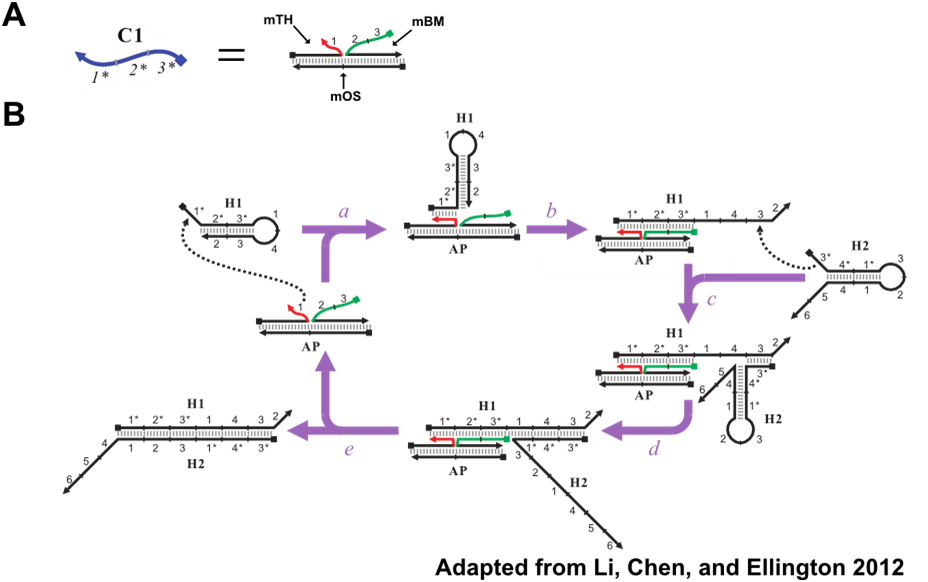
Further Modifications of the Catalyst
The scCHA catalyst can be further modified in order to detect minor structural defects in mOS. By including limited complementarity, 1-2 bp, between the 1 domain of mTH and mOS, the rate of catalysis can be greatly reduced, Figure 9. When the 1 domain is partially annealed to mOS it is hindered from annealing to the toehold on H1. However, if a defect is present in mOS in this region of complementarity, the 1 domain will be freed to function normally and the rate of catalysis is restored. For our purposes this defect can be an SNP allele or an AP site. mOS is the target sequence to be detected and characterized by our circuit.

MDM2 Circuit Design
In order to have a circuit that would specifically detect and characterize SNP309 of the MDM2 gene, new CHA circuit pieces were designed using CircDesigNA. Hairpins were designed to not interact with the MDM2 gene in the absence of the split catalyst pieces, Table 1. The split catalyst was designed as depicted in Figure 10. The MDM2 gene sequence determined a majority of the split catalyst sequences and the remaining sequences were designed by CircDesigNA.

| Oligo Name | Description | Sequence (5' to 3') |
|---|---|---|
| MDM2 | ||
| BioM_MDM2_Cat_1,2,3p1 | Continuous catalyst for testing circuit hairpins | CTTCAAACCCTAAATCGACACAAC |
| BioM_MDM2_Cat_1p1 | Split catalyst piece containing domain 1 | CACCTGCGATCATCCGGACCTCCCGCGCCGACACAAC |
| BioM_MDM2_Cat_2,3p1 | Split catalyst piece containing domains 2 and 3 | CTTCAAACCCTAAATCGCGGCCCCGCAGCCCCCGGCCCCCGTGAC |
| BioM_MDM2_H1p1 | Hairpin 1 for MDM2 detection and characterization | GTTGTGTCGATTTAGGGTTTGAAGCTCTCTCCCTTCAAACCCTAAATCCCTCCCTCCCTCCCTC |
| BioM_MDM2_H2p1 | Hairpin 2 for MDM2 detection and characterization | GTTTGAAGGGAGAGAGCTTCAAACCCTAAATCCTCTCTCC |
| BioM_MDM2_RepFp1 | Flurophore tagged reporter oligo for MDM2 detection and characterization | /56-FAM/GAGGGAGGGAGGGAGGGATTTAGG |
| BioM_MDM2_RepQp1 | Quencher tagged reporter oligo for MDM2 detection and characterization | CCTCCCTCCCTCCCTC/3IABkFQ/ |
| Apurinic Site | ||
| BioM_Cat_1,2,3p1 | Continuous catalyst for testing circuit hairpins | CTTTTCTGCATCTATCTCCTAACC |
| BioM_Cat_1p1 | Split catalyst piece containing domain 1 | GAACCCCCGGTTCCTCTCCTAACC |
| BioM_Cat_2,3p1 | Split catalyst piece containing domains 2 and 3 | CTTTTCTGCATCTATCGTACTGAGGGGGCGTA |
| BioM_H1p1 | Hairpin 1 for AP site detection and characterization | GGTTAGGAGATAGATGCAGAAAAGCAATTGTCCTTTTCTGCATCTATCGAAGTAAGGTAGTGTG |
| BioM_H2p1 | Hairpin 2 for AP site detection and characterization | CAGAAAAGGACAATTGCTTTTCTGCATCTATCCAATTGTC |
| BioM_RepFp1 | Flurophore tagged reporter oligo for AP site detection and characterization | /56-FAM/GTAGTGTGGAAGTAAGGATAGATG |
| BioM_RepQp1 | Quencher tagged reporter oligo for AP site detection and characterization | CTTACTTCCACACTAC/3IABkFQ/ |
Apurinic Circuit Design
The apurinic circuit was designed in the same fashion as the MDM2 circuit. The catalyst pieces were designed to assemble on the A4324 loop of the 28S rRNA, Figure 11. The remaining circuit pieces for the apurinic design can be found in Table 1.
