Biomod/2013/UT-Austin: Difference between revisions
No edit summary |
No edit summary |
||
| Line 141: | Line 141: | ||
====Page Purification Protocol==== | ====Page Purification Protocol==== | ||
Revision as of 12:24, 26 October 2013
Introduction
Modern medicine is defined by it's relationship with large centralized medical laboratories. Advances in biomedical science has led to the development of many powerful, yet expertise and resource intensive, diagnostic techniques. Due to these expertise and resource requirements, the deployment of these modern techniques was realized through the development of large centralized medical laboratories. These labs have greatly expanded a physician’s ability to diagnose, and in turn, treat patients. However, these labs are not without their flaws. Testing through a central laboratory increases the time between attainment of the patient’s fluid or tissue sample to delivery of test results to the physician. Additionally, the current state of dependence upon these labs has hindered the deployment of modern care to resource poor areas.
Point-of-Care Diagnostics and Biosensors
Point-of-care (POC) diagnostics are diagnostic technologies that move the analysis of patient samples from the lab bench to the bedside and in some cases the home. More specifically, POC diagnostics are small and simple to operate devices that can process complex clinical samples for various biomarkers. This both decreases the time to diagnosis and removes some of the barriers to deployment of modern care into the field.

Frequently, the miniaturization and simplification of sample analysis is achieved through the use of biosensors. An illustrative example of this is the home pregnancy test. Modern pregnancy tests use antibodies to detect elevated levels of hormones during pregnancy. Specifically, most pregnancy test use anti-human chorionic gonadotropin (hCG) conjugated with colloidal metals to visualize elevated hCG in the urine of pregnant women.
Proteins vs Nucleic Acids As Biosensors
While modern pregnancy tests are relatively rugged, i.e. no refrigeration or exhaustive storage criteria exist, POC devices based on protein components, the development of protein-based biosensors is frequently hindered by the inherent instability of protein components. Various techniques for stabilization of useful proteins, e.g. antibodies and enzymes, are in development. However, the tendency of proteins to irreversibly denature remains highly problematic.
In this context, functional nucleic acids offer the potential to replace proteins in the development of rugged biosensors. Nucleic acids (NAs) can generally be reversibly denatured and, when desiccated and isolated from nucleases, degrade on a millennial timescale. NAs are amenable to a variety of conjugation chemistries that allow for attachment of a range of functional groups and dyes. Additionally NAs, like proteins, can specifically recognize biomarkers through both formation of complex tertiary structure, e.g. aptamers, and, in the case of genomic material, direct hybridization. The predictable nature of these hybridization interactions has enabled the development of complex DNA circuits for both biomarker signal amplification and transduction.
DNA Circuitry
Toehold-Mediated Strand Displacement
Toehold-mediated strand displacement is a specific hybridization interaction that is frequently exploited for the construction of complex DNA circuits. The process, illustrated in Figure 2, begins with an ssDNA oligo, green, binding a short, frequently 4-10 base pairs in length, single stranded domain of a DNA duplex, red and blue. This single stranded domain is deemed a toehold, as it allows the green oligo to weakly associate with the duplex through hybridization to this small single stranded domain. The green oligo then undergoes a process referred to as branch migration. Branch migration is the random walk process in which one domain displaces another of identical sequence through a series of reversible single nucleotide dissociation and hybridization steps. While each hybridization event in branch migration is reversible, the greater total complementarity of the green strand to the blue strand makes displacement of the red strand thermodynamically favorable.

Catalyzed Hairpin Assembly
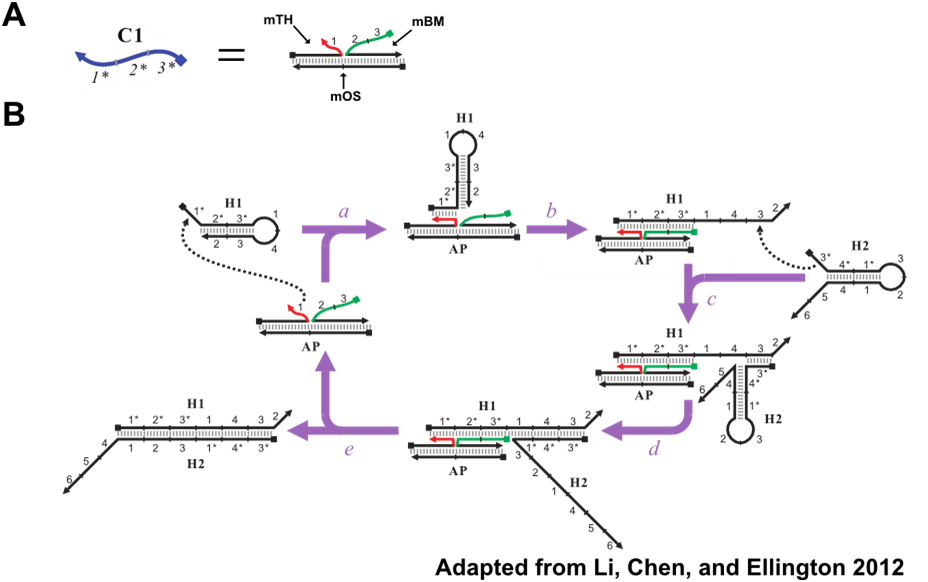
Catalyzed hairpin assembly (CHA) is an enzyme-free nucleic acid signal amplification circuit that enables specific detection of oligonucleotides. The signal amplification reaction involves three oligos, depicted in Figure 3: the catalyst (C1), the first hairpin (H1), and the second hairpin (H2). The reaction begins with C1 opening the H1 hairpin by toehold-mediated strand displacement through the 1 toehold of H1. The now linear H1 now has an exposed 3* domain. This acts as a toehold for H2, in turn, allowing H2 to fully hybridize to H1. C1, which is only weekly associated with the H1:H2 duplex, will eventually dissociate, thereby completing one cycle of the reaction and freeing C1 to generate additional H1:H2 duplexes. In this fashion, the presence, or signal, of a few catalyst molecules is amplified through the generation of numerous H1:H2.

In addition to amplifying signal, the formation of H1:H2 fully exposes the 2*, 5* and 6* domains, in red, of H1. These domains can be modified and used in concert with reporter oligonucleotides to produce various forms of signals for the presence of H1:H2. Figure 4 depicts CHA reporter schemes that include fluorescent, colorimetric, and electrochemical readout. All reporter schemes make use of the 2* domain that is exposed on formation of H1:H2.

The potential impact of CHA as an analytical tool inspired our team to investigate if CHA could not only detect oligonucleotides but also characterizes them. With slight design modifications, discussed in the Design section, CHA can detect small point defects in oligos. The ability to characterize oligos with an enzyme free circuit would potentially allow for the development of rugged POC diagnostic devices for detection of genetic abnormalities, virulent strains of bacteria, and the activity of toxins that act upon nucleic acids. Therefore, our team decided to develop modified CHA circuits for the detection of cancer-linked MDM2 single nucleotide polymorphisms (SNPs) and ricin toxin generated apurinic sites.
Single Nucleotide Polymorphisms
A single nucleotide polymorphism (SNP) is a DNA sequence variation at a single position, i.e. an A, T, C, or G, within a population. SNPs account for a large portion of human genetic diversity. For example, twenty-five percent of the population may possess an A-T pair at a certain locus, while the remaining seventy-five percent possess a G-C pair. In the case of human SNPs, certain SNP alleles have been associated with adverse responses to certain medications, susceptibility to infectious diseases, and propensity to develop cancer. In the case of bacterial SNPs, certain alleles have been associated with heightened virulence or resistance to antibiotics.
MDM2 SNP 309
Our specific target for this project is SNP309 of the mouse double minute 2 homolog (MDM2) gene. This is a well characterized SNP that is associated with increase cancer risk. SNP 309, located in the promoter of the gene, is a T/G SNP with the G allele being the cancer-linked allele. The G allele increases cancer risk by up regulating expression of MDM2. This up regulation leads to increased cancer risk due to MDM2’s ability to binds to and inhibits the tumor suppressor protein, p53, depicted in Figure 5. Therefore, easy detection of SNP309 would be useful for evaluating an individual’s cancer risk.

Current SNP Detection Methods
The most common method of SNP detection is real-time PCR (rtPCR). rtPCR is essentially PCR with fluorescent probes, depicted in Figure 6, to quantify the products of the reaction. Probe designs vary but all operate by exploiting the single base pair mismatch that can result from SNP allele variation.

Apurinic Sites
Similar to SNPs, apurinic (AP) sites affect a single nucleotide. Apurinic sites, depicted in Figure 7, occur when the purine ring is removed from the ribose group of the nucleic acid backbone. For design purposes, apurinic sites should behave in a similar fashion to single base pair mismatches associated with SNPs.
Ricin Toxin Generated AP Sites
Ricin toxin, found in the seeds of the castor oil plant, is a highly lethal toxin that acts by inactivating the ribosome. Specifically, ricin toxin depurinates the 4324 adenine of the 28S ribosomal RNA, depicted in Figure 7. This depurination weakens the binding of elongation factors to the A4324 loop and prevents its function. Without functioning elongation factors, the ribosome cannot translocate tRNAs following peptidyl transfer. The inability to produce new proteins leads to rapid cell death.

Current Ricin Detection Methods
Current ricin toxin detection methods are relatively limited. In vivo, lethal dose delivered to mice, assays and in situ, cytotoxicity, assays exist. However, recent efforts have been focused on attaining in vitro assays for ricin activity. These assays would allow for easier more rapid detection of ricin.
Design
The CHA reaction described in the introduction allows for the detection of oligonucleotides. The oligo detected is C1, Figure 8A. C1 possesses 3 domains: the toehold binding domain (1*) and the domains that undergo branch migration in order to open H1. However, previous work (Li, Chen, and Ellington 2012) has shown that these catalytic domains do not have to be found on a single oligo. These domains can be split between separate oligos and still trigger CHA when colocalized on a common template, Figure 8B. This modified variant of CHA will be referred to as split catalyst CHA (scCHA) in this work.
Further Modifications of the Catalyst
The scCHA catalyst can be further modified in order to detect minor structural defects in mOS. By including limited complementarity, 1-2 bp, between the 1 domain of mTH and mOS, the rate of catalysis can be greatly reduced, Figure 9. When the 1 domain is partially annealed to mOS it is hindered from annealing to the toehold on H1. However, if the a defect is present in mOS in this region of complementarity, the 1 domain will be freed to function normally, in turn, restoring the rate of catalysis. For our purposes, this defect can be an SNP allele or an apurinic site. mOS is the target sequence to be detected and characterized by our circuit.

MDM2 Circuit Design
In order to design a circuit that would specifically detect and characterize SNP309 of the MDM2 gene, new CHA circuit pieces were designed using CircDesigNA. Hairpins were designed as to not interact with MDM2 DNA in the absence of the split catalyst pieces, Table 1. The split catalyst was designed as depicted in Figure 10. The MDM2 gene sequence determined a majority of the split catalyst sequence. The remaining sequence was determined by CircDesigNA.

| Oligo Name | Description | Sequence |
|---|---|---|
| MDM2 | ||
| BioM_MDM2_Cat_1,2,3p1 | Continuous catalyst for testing circuit hairpins | CTTCAAACCCTAAATCGACACAAC |
| BioM_MDM2_Cat_1p1 | Split catalyst piece containing domain 1 | CACCTGCGATCATCCGGACCTCCCGCGCCGACACAAC |
| BioM_MDM2_Cat_2,3p1 | Split catalyst piece containing domains 2 and 3 | CTTCAAACCCTAAATCGCGGCCCCGCAGCCCCCGGCCCCCGTGAC |
| BioM_MDM2_H1p1 | Hairpin 1 for MDM2 detection and characterization | GTTGTGTCGATTTAGGGTTTGAAGCTCTCTCCCTTCAAACC
CTAAATCCCTCCCTCCCTCCCTC |
| BioM_MDM2_H2p1 | Hairpin 2 for MDM2 detection and characterization | GTTTGAAGGGAGAGAGCTTCAAACCCTAAATCCTCTCTCC |
| BioM_MDM2_RepFp1 | Flurophore tagged reporter oligo for MDM2 detection and characterization | /56-FAM/GAGGGAGGGAGGGAGGGATTTAGG |
| BioM_MDM2_RepQp1 | Quencher tagged reporter oligo for MDM2 detection and characterization | CCTCCCTCCCTCCCTC/3IABkFQ/ |
| Apurinic Site | ||
| BioM_Cat_1,2,3p1 | Continuous catalyst for testing circuit hairpins | CTTTTCTGCATCTATCTCCTAACC |
| BioM_Cat_1p1 | Split catalyst piece containing domain 1 | GAACCCCCGGTTCCTCTCCTAACC |
| BioM_Cat_2,3p1 | Split catalyst piece containing domains 2 and 3 | CTTTTCTGCATCTATCGTACTGAGGGGGCGTA |
| BioM_H1p1 | Hairpin 1 for apurinic site detection and characterization | GGTTAGGAGATAGATGCAGAAAAGCAATTGTCCTTTTCT
GCATCTATCGAAGTAAGGTAGTGTG |
| BioM_H2p1 | Hairpin 2 for apurinic site detection and characterization | CAGAAAAGGACAATTGCTTTTCTGCATCTATCCAATTGTC |
| BioM_RepFp1 | Flurophore tagged reporter oligo for apurinic site detection and characterization | /56-FAM/GTAGTGTGGAAGTAAGGATAGATG |
| BioM_RepQp1 | Quencher tagged reporter oligo for apurinic site detection and characterization | CTTACTTCCACACTAC/3IABkFQ/ |
Apurinic Circuit Design
The apurinic circuit was designed in the same fashion as the MDM2 circuit. However, the catalyst pieces assembled on the A4324 loop of the 28S rRNA, Figure 11. The remaining circuit pieces for the apurinic design can be found in Table 1.

Methods
Computational
CircDesigNA
CircDesigNA is nucleic acid design software that interfaces with NUPACK to generate sequence for the design of oligonucleotides with a desired secondary structure. The user can specify complementary domains using the nomenclature presented in the circuit figures. Additionally, users can specify sequence constraints, such as those dictated by our target sequences.
NUPACK
NUPACK is a free web-based software developed by Caltech, and is useful for analyzing nucleic acid systems. This program provides information regarding the most thermodynamically stable nucleic acid structures, specifically for DNA to DNA hybridization and RNA to RNA hybridization. There is no information provided for DNA to RNA hybridization.
Geneious
Geneious is a bioinformatics software program. For our purposes it was useful for designing and annotating the MDM2 gene except construct.
OligoAnalyzer
The OligoAnalyzer on the IDT website was used to estimate primer melting temperatures during PCR.
Primer Design
Primers were designed using Geneious. Melting temperatures were estimated using IDT’s OligoAnalyzer and unwanted secondary structure was avoided by NUPACK analysis.
Experimental
Oligonucleotide Preparation
All oligonucleotides were ordered from Integrated DNA Technologies (Coralville, IA). Non-HPLC purified oligos were purified through polyacrylamide gel electrophoresis (PAGE) and concentrated by ethanol precipitation.