Biomod/2013/StJohns/results: Difference between revisions
No edit summary |
|||
| (35 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
*We have synthesized versions of the claw with (‘sticky’) and without (‘blunt’) single-stranded binding elements. | *We have synthesized versions of the claw with (‘sticky’) and without (‘blunt’) single-stranded binding elements. | ||
*We have synthesized versions of the claw with and without fluorescent tags for FRET analysis. | *We have synthesized versions of the claw with and without fluorescent tags for FRET analysis. | ||
*We have visualized the above versions of the claw on AFM to show that they take the predicted shape. | *[[Biomod/2013/StJohns/results#Claw Visualization|We have visualized the above versions of the claw on AFM to show that they take the predicted shape.]] | ||
*We have shown that the above versions of the claw form tight bands on a gel, indicating a single primary product of the anneal. | *We have shown that the above versions of the claw form tight bands on a gel, indicating a single primary product of the anneal. | ||
*We have demonstrated a binding interaction between the functionalized claw and functionalized capsid as well as a lack of interaction between the nonfunctionalized claw and capsid. | *[[Biomod/2013/StJohns/results#FRET|We have verified that FRET-tagged origami can be visualized in a gel, providing easy visual discrimination between differently-tagged versions of the claw.]] | ||
*We have not been able to differentiate bound and unbound complexes via DLS. | *'''We have not successfully shown FRET phenomena involving the bound claw-capsid complex.''' | ||
*We have generated and isolated FAB fragments for use as binding elements in future claw designs. | *[[Biomod/2013/StJohns/results#Binding|We have demonstrated a binding interaction between the functionalized claw and functionalized capsid as well as a lack of interaction between the nonfunctionalized claw and capsid, and explored the anneal conditions to optimize the interaction.]] | ||
*We have demonstrated the potential for selecting claws based on their avidity using a chromatography augmented with photocleavable elements. | *[[Biomod/2013/StJohns/results#DLS|'''We have not been able to differentiate bound and unbound complexes via DLS.''']] | ||
*[[Biomod/2013/StJohns/results#FAB|We have generated and isolated FAB fragments for use as binding elements in future claw designs.]] | |||
*'''We have not been able to effectively conjugate purified DNA strands to our ketone models for FAB incorporation.''' | |||
*[[Biomod/2013/StJohns/results#Selection|We have demonstrated the potential for selecting claws based on their avidity using a chromatography augmented with photocleavable elements.]] | |||
*[[Biomod/2013/StJohns/results#Triangles|We have synthesized and characterized DO triangles with precisely controlled vertex angles.]] | |||
=Data= | |||
==Claw Visualization== | |||
Below are AFM images of the claws alone, with and without binding elements. | |||
[[Image:Lukemanlab-2013-0020.jpg|frameless|center|400px]] | |||
==Binding== | |||
===Initial results=== | |||
We used gel electrophoresis to characterize the binding interaction between origami structures and capsids with and without binding elements. | |||
[[Image:Lukemanlab-2013-0001.png|thumb|400px|center|No binding between WT capsids and any DO,<br> | [[Image:Lukemanlab-2013-0001.png|thumb|400px|center|No binding between WT capsids and any DO,<br> | ||
| Line 21: | Line 29: | ||
however, blunt DO binds sticky capsids.]] | however, blunt DO binds sticky capsids.]] | ||
===Stoichiometric control=== | |||
We have also demonstrated that the band shift on a gel is dependent on the stoichiometry of the binding anneal, implying that this technique can be used to detect not only presence-absence of the target substrate but also the concentration of the substrate. | |||
[[Image:Lukemanlab-2013-0018.jpg|frameless|400px|center]] | |||
===Optimization=== | |||
After exploring a number of avenues for optimizing binding conditions, we've managed to increase the specificity of the binding, thus improving our signal-to-noise ratio for systems based on this design. Tested factors include gel temperature, annealing at various constant temperatures and across various temperature gradients for different lengths of time, varied concentration of Mg<sup>2+</sup> ions in buffer, addition of T15 in various ratios, 10% and 20% formamide in buffer solution, and short (TCA)4 and (TCA)5 "Nanny" strands. | |||
[[Image:Lukemanlab-2013-0014.png|frameless|400px|center|Lanes, left to right:<br> | |||
Ladder, wt, T21, BC, BC + wt, BC + T21, SC, SC + wt, SC + T21<br> | |||
wt: unmodified capsid<br> | |||
T21: Functionalized capsid / target substrate<br> | |||
BC: Unmodified, "blunt" claw | |||
SC: Functionalized, "sticky" claw]] | |||
We are currently searching for conditions to improve the signal-to-noise ratio further. | |||
==FRET== | |||
Below is a gel demonstrating that versions of the claw tagged with different fluorescent elements can be visually distinguished on a gel. | |||
[[Image:Lukemanlab-2013-0002.png|thumb|400px|center|Each different color band corresponds to a different fluorescent tag.<br>The six bands on the left are 'blunt' claw and the six on the right are 'sticky' claw.<br>Odd-numbered lanes contain 0.1 pmol claw, while even-numbered lanes contain 0.2 pmol.]] | |||
==DLS== | |||
After optimising laser conditions to work with these dilute nanomolar solutions of origami and capsid, we managed to show that the origami control structure and the capsid are of different sizes. However, when performing DLS on a mix of nonfunctionalized origami and capsid, we found only a single peak rather than peaks showing two differently-sized objects. Based on [[Biomod/2013/StJohns/results#Binding Interaction|data from other experiments]], it is unlikely that this represents a binding interaction. We are currently investigating the source of this anomaly. | |||
[[Image:Lukemanlab-2013-0003.png|thumb|400px|center]] | |||
==Selection== | |||
[[Image:Lukemanlab-2013-0004.jpg|thumb|400px|center| The blue boxes highlight where bands would be expected if the photocleaving reaction failed. <br> Note the presence of only functionalized claw in the post-cleavage solution.]] | |||
==FAB== | |||
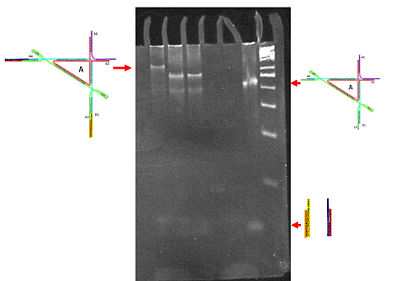
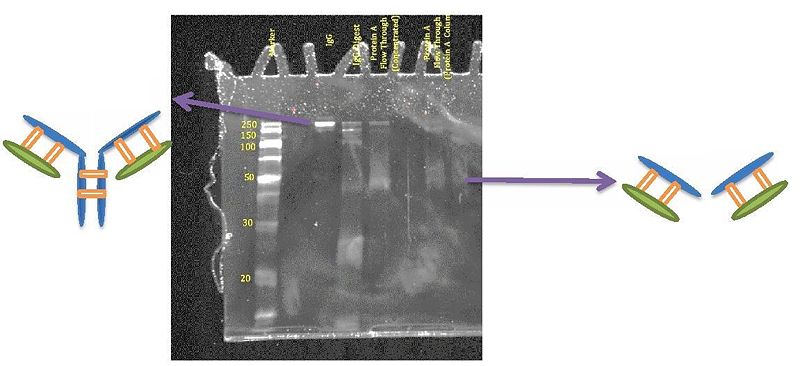
We made many attempts at the literature reported papain digestion of IgG into Fab fragments without success. We switched to another enzyme system. Gratifyingly, using Ficin, we obtained Fab fragments. The following are the gel results obtained for the IgG sample. | |||
Notice that in the first gel, the IgG shows up at around 150 – 250 kDa which agrees with what one would expect. Moreover, in the IgG digest, the band that is close to 50 kDa is the product of interest the Fab fragments. But notice that there are a lot of other impurities that interfere with the sample below 50 kDa. In order to get rid of all those bands, a purification step is needed. | |||
[[Image:Lukemanlab-2013-0005.jpg|frameless|800px|center]] | |||
Notice that after purifying the samples with the 30K spin columns, there are no bands below 30 kDa. This shows that the purification step was successful. | |||
[[Image:Lukemanlab-2013-0017.jpg|frameless|800px|center]] | |||
==Triangles== | |||
Utilizing atomic force microscopy (AFM), we managed to image 90 degree origami triangles with two groups of ‘extenders’. These ‘extenders’ are duplexes that have been made at precise stoichiometric ratios. The goal of this project was to successfully attach these two duplexes to the sticky ends of these 90 degree triangles. After obtaining several AFM images showing what appeared to be several triangles with duplexes attached to them, this AFM picture shown here demonstrates the best resolution and separation between each respective triangle. The duplexes that attached to the triangles had a complementary sequence to the sticky ends on the triangle, which is what made it possible for attachment. We have had difficulty in achieving a constant 90 degree angle with both of the duplexes attached. However, when measuring the angles of the triangles with both groups of ‘extenders’ attached, most of our angles were precise (around 100 degrees). In future projects, we hope to gather enough images (such as these) to plot the different angles obtained in an attempt to get as close as possible to 90 degrees. | |||
The | |||
[[Image:Lukemanlab-2013-0009.tif|frameless|400px|center]] | |||
In examining these structures via gel electrophoresis, we would expect that the 90 degree origami triangle with both of its duplexes (or extenders) would run at a slower mobility than the triangle by itself or with one pair of duplexes. It became clear that this sample of triangle with both duplexes (which is the same sample that was used to generate the AFM image above) was a completely different specimen as compared to the other DNA complexes. We utilized a 50 base pair DNA marker to compare the different sizes of the complexes. What is most exciting is that we obtained a sharp and tight band meaning that our sample was one that had a specific DNA complex configuration. This was further supported in our generated AFM images. In future experiments, we could attempt to run a gel similar to this one at either a lower concentration or higher temperatures to eliminate the accumulation of oligomers in the wells. | |||
[[Image:Lukemanlab-2013-0010.jpg|frameless|400px|center]] | |||
Latest revision as of 17:11, 26 October 2013
<html>
<head>
<link href='http://fonts.googleapis.com/css?family=Open+Sans' rel='stylesheet' type='text/css'>
</head>
<style>
body {
font-family: 'Trebuchet MS', 'Open Sans', sans-serif; overflow-y: auto;
}
.container {
background-color: #ffffff; margin-top:0px
} .OWWNBcpCurrentDateFilled { display: none; }
h5 {
font-family: 'Trebuchet MS', 'Open Sans', sans-serif; font-size: 11px; font-style: normal; text-align: center; margin:0px; padding:0px;
}
- column-content
{
width: 0px; float: left; margin: 0 0 0 0; padding: 0;
} .firstHeading {
display:none; width:0px;
}
- column-one
{
display:none; width:0px; background-color: #ffffff;
}
- globalWrapper
{
width: 1024px; background-color: #ffffff; margin-left: auto; margin-right: auto
}
- content
{
margin: 0 0 0 0; align: center; padding: 12px 12px 12px 12px; width: 1000px; background-color: #ffffff; border: 0;
}
- bodyContent
{
width: 974px; align: justify; background-color: #fffffff;
}
- column-content
{
width: 1024px; background-color: #ffffff;
}
- footer
{
position: absolute; bottom: 0; width: 1024px;
}
- menu
{
position: fixed; float: left; width: 180px; padding: 10px; background-color: #FFFFFF;
}
- resolutionwarning
{
display: none; text-align: center;
}
- pagecontent
{
text-align: justify; float: right; width: 744px; margin-left: 300px; min-height: 400px
}
- toc { display: none; }
@media all {
body { background: #ffffff 0 0 no-repeat;}
}
/*Expanding list*/ ul { list-style: none; }
- exp li ul { display: none; }
- exp li:hover ul { display: block; }
- exp li a:active ul { display: block; }
a:link {color:#007788;} /*unvisited link*/ a:visited {color:#007788;} /* visited link */ a:hover {color:#00ccff;} /* mouse over link */ a:active {color:#00ccff;} /* selected link */
</style> </html>
Summary
- We have synthesized versions of the claw with (‘sticky’) and without (‘blunt’) single-stranded binding elements.
- We have synthesized versions of the claw with and without fluorescent tags for FRET analysis.
- We have visualized the above versions of the claw on AFM to show that they take the predicted shape.
- We have shown that the above versions of the claw form tight bands on a gel, indicating a single primary product of the anneal.
- We have verified that FRET-tagged origami can be visualized in a gel, providing easy visual discrimination between differently-tagged versions of the claw.
- We have not successfully shown FRET phenomena involving the bound claw-capsid complex.
- We have demonstrated a binding interaction between the functionalized claw and functionalized capsid as well as a lack of interaction between the nonfunctionalized claw and capsid, and explored the anneal conditions to optimize the interaction.
- We have not been able to differentiate bound and unbound complexes via DLS.
- We have generated and isolated FAB fragments for use as binding elements in future claw designs.
- We have not been able to effectively conjugate purified DNA strands to our ketone models for FAB incorporation.
- We have demonstrated the potential for selecting claws based on their avidity using a chromatography augmented with photocleavable elements.
- We have synthesized and characterized DO triangles with precisely controlled vertex angles.
Data
Claw Visualization
Below are AFM images of the claws alone, with and without binding elements.

Binding
Initial results
We used gel electrophoresis to characterize the binding interaction between origami structures and capsids with and without binding elements.

‘sticky’ DO bind sticky capsids strongly;
however, blunt DO binds sticky capsids.
Stoichiometric control
We have also demonstrated that the band shift on a gel is dependent on the stoichiometry of the binding anneal, implying that this technique can be used to detect not only presence-absence of the target substrate but also the concentration of the substrate.

Optimization
After exploring a number of avenues for optimizing binding conditions, we've managed to increase the specificity of the binding, thus improving our signal-to-noise ratio for systems based on this design. Tested factors include gel temperature, annealing at various constant temperatures and across various temperature gradients for different lengths of time, varied concentration of Mg2+ ions in buffer, addition of T15 in various ratios, 10% and 20% formamide in buffer solution, and short (TCA)4 and (TCA)5 "Nanny" strands.

Ladder, wt, T21, BC, BC + wt, BC + T21, SC, SC + wt, SC + T21
wt: unmodified capsid
T21: Functionalized capsid / target substrate
BC: Unmodified, "blunt" claw SC: Functionalized, "sticky" claw
We are currently searching for conditions to improve the signal-to-noise ratio further.
FRET
Below is a gel demonstrating that versions of the claw tagged with different fluorescent elements can be visually distinguished on a gel.

The six bands on the left are 'blunt' claw and the six on the right are 'sticky' claw.
Odd-numbered lanes contain 0.1 pmol claw, while even-numbered lanes contain 0.2 pmol.
DLS
After optimising laser conditions to work with these dilute nanomolar solutions of origami and capsid, we managed to show that the origami control structure and the capsid are of different sizes. However, when performing DLS on a mix of nonfunctionalized origami and capsid, we found only a single peak rather than peaks showing two differently-sized objects. Based on data from other experiments, it is unlikely that this represents a binding interaction. We are currently investigating the source of this anomaly.

Selection

Note the presence of only functionalized claw in the post-cleavage solution.
FAB
We made many attempts at the literature reported papain digestion of IgG into Fab fragments without success. We switched to another enzyme system. Gratifyingly, using Ficin, we obtained Fab fragments. The following are the gel results obtained for the IgG sample.
Notice that in the first gel, the IgG shows up at around 150 – 250 kDa which agrees with what one would expect. Moreover, in the IgG digest, the band that is close to 50 kDa is the product of interest the Fab fragments. But notice that there are a lot of other impurities that interfere with the sample below 50 kDa. In order to get rid of all those bands, a purification step is needed.

Notice that after purifying the samples with the 30K spin columns, there are no bands below 30 kDa. This shows that the purification step was successful.
Triangles
Utilizing atomic force microscopy (AFM), we managed to image 90 degree origami triangles with two groups of ‘extenders’. These ‘extenders’ are duplexes that have been made at precise stoichiometric ratios. The goal of this project was to successfully attach these two duplexes to the sticky ends of these 90 degree triangles. After obtaining several AFM images showing what appeared to be several triangles with duplexes attached to them, this AFM picture shown here demonstrates the best resolution and separation between each respective triangle. The duplexes that attached to the triangles had a complementary sequence to the sticky ends on the triangle, which is what made it possible for attachment. We have had difficulty in achieving a constant 90 degree angle with both of the duplexes attached. However, when measuring the angles of the triangles with both groups of ‘extenders’ attached, most of our angles were precise (around 100 degrees). In future projects, we hope to gather enough images (such as these) to plot the different angles obtained in an attempt to get as close as possible to 90 degrees.

In examining these structures via gel electrophoresis, we would expect that the 90 degree origami triangle with both of its duplexes (or extenders) would run at a slower mobility than the triangle by itself or with one pair of duplexes. It became clear that this sample of triangle with both duplexes (which is the same sample that was used to generate the AFM image above) was a completely different specimen as compared to the other DNA complexes. We utilized a 50 base pair DNA marker to compare the different sizes of the complexes. What is most exciting is that we obtained a sharp and tight band meaning that our sample was one that had a specific DNA complex configuration. This was further supported in our generated AFM images. In future experiments, we could attempt to run a gel similar to this one at either a lower concentration or higher temperatures to eliminate the accumulation of oligomers in the wells.