Biomod/2013/Aarhus/Supplementary/Supplementary Data
<html> <style> /* ul.menu li.</html>Supplementary/Supplementary Data<html> a {
color: cyan;
}
- /
- toc {
display: none; }
- mytoc {
background: none; width: 200px; }
.toc { border: 0px solid; }
- toc ul ul,.toc ul ul {
margin: 0 0 0 1em; }
table.toc { background-color: #f0f4f4; }
- mytoc a,#mytoc a:visited {
font-size: normal; color: #222222; }
- mytoc a:hover {
font-color: #009ee0; /* text-decoration: underline; */ }
- wiki-toc {
width: 200px; margin-top: 6px; float: left; }
- wiki-body {
margin-left: 200px; padding-left: 12px; padding-right: 35px; }
- toc #toctitle,.toc #toctitle,#toc .toctitle,.toc .toctitle {
text-align: left; }
- toc h2,.toc h2 {
font-weight: normal; font-size: 17px; }
/*
- wiki-contents A {
color: #00aeef; }
- wiki-contents A:HOVER {
color: #00aeef; }
- /
- toctitle span {
display: none; }
/*
- wiki-body p,#wiki-body li,#wiki-body dd,div.thumbcaption {
font-size: medium; }
- /
/* required to avoid jumping */
- tocScrolWrapper {
/* left: 450px; */ position: absolute; /* margin-left: 35px; width: 280px; */ }
- tocScrol {
position: absolute; top: 0; /* just used to show how to include the margin in the effect */ /*margin-top: 20px; */ /* border-top: 1px solid purple; */ /*padding-top: 19px;*/ }
- tocScrol.fixed {
position: fixed; top: 0; }
- editPageTxt {
text-align: left; padding-left: 15px; }
- editPageTxt P {
clear: both; }
- toPageTop {
float: left; position: relative; top: 18px; left: 13px; color: #d13f31; } </style>
<script type="text/javascript"> $(document).ready(function() { var parentTable = $("#toc").parent(); $('#mytoc').append($("#toc").first());
$('#mytoc').find("#toc").attr("id", ""); parentTable.closest('table').remove(); });
$(document).ready( function() { var top = $('#tocScrol').offset().top - parseFloat($('#tocScrol').css('marginTop').replace( /auto/, 0)); var nav = $('#tocScrol'); var max = $('#indexing').offset().top - nav.height();
$(window).scroll(function(event) { // what the y position of the scroll is var y = $(this).scrollTop();
if (y > top) { // && signs are html decoded thus this construction if (y >= max) { nav.removeClass('fixed'); nav.css({ position : 'absolute', top : max - top }); } else { nav.addClass('fixed'); nav.removeAttr('style'); } } else { nav.removeClass('fixed'); nav.removeAttr('style'); } }); }); </script> <html> <html> <div id="wiki-contents"> <div id="tocScrolWrapper"> <div id="tocScrol"> <div id="wiki-toc"> <a id="toPageTop" href="#">▲</a> <table id="mytoc" class="toc" summary="Contents"> </table> <div id="editPageTxt"> <p> [<a href="http://openwetware.org/index.php?title=Biomod/2013/Aarhus/</html>Supplementary/Supplementary Data<html>&action=edit">edit this page</a>] </p> </div> </div> </div> </div> <div id="wiki-body"> </html>
Supplementary data
Origami design



sisiRNA
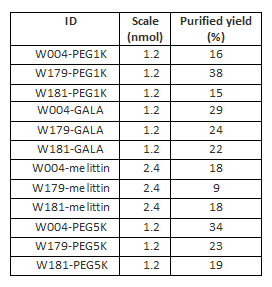
Yields of RP-HPLC purifications of the modified strands
Estimated yields of the annealing reactions

Luciferase knockdown with all singly modified duplexes

System in action
Radioactive labeling of W376

The band containing the labeled sample appears most similar to that in lane 4 which means that the 6 µL that were run on the gel contained 1.5 pmol W376, corresponding to a concentration of 0.25 µM in the purified sample.
Cholesterol
1H NMR, 13C NMR and IR of compound 4.
<html><A name="NMR4H"/></html>

<html><A name="NMR4C"/></html>


<html><A name="IR4"/></html>

1H NMR, 13C NMR and IR of compound 5.
<html><A name="NMR5H"/></html>

<html><A name="NMR5C"/></html>


<html><A name="IR5"/></html>

1H NMR and 13C NMR of compound 6.
<html><A name="NMR6H"/></html>

<html><A name="NMR6C"/></html>


Photosensitizer
1H NMR and 13C NMR of compound 7.
<html><A name="NMR7H"/></html>

<html><A name="NMR7C"/></html>

1H NMR and 13C NMR of compound 8.
<html><A name="NMR8H"/></html>

<html><A name="NMR8C"/></html>

1H NMR, 13C NMR and UV of compound 9.
<html><A name="NMR9H"/></html>

<html><A name="NMR9C"/></html>

1H NMR, 13C NMR and UV of compound 10.
<html><A name="NMR10H"/></html>

<html><A name="NMR10C"/></html>

1H NMR and 13C NMR of compound 11.
<html><A name="NMR11H"/></html>

<html><A name="NMR11C"/></html>

1H NMR and 13C NMR of compound 12.
<html><A name="NMR12H"/></html>

<html><A name="NMR12C"/></html>

5-propargylamino-ddUTP
1H NMR, 13C NMR and IR of compound 15.
<html><A name="NMR15H"/></html>


<html><A name="NMR15C"/></html>

<html><A name="IR15"/></html>

1H NMR, 13C NMR and IR of compound 16.
<html><A name="NMR16H"/></html>


<html><A name="NMR16C"/></html>

<html><A name="IR16"/></html>

1H NMR, 13C NMR and IR of compound 17.
<html><A name="NMR17H"/></html>


<html><A name="NMR17C"/></html>

<html><A name="IR17"/></html>

1H NMR, 13C NMR and IR of compound 18.
<html><A name="NMR18H"/></html>


<html><A name="NMR18C"/></html>

<html><A name="IR18"/></html>

1H NMR and 19F NMR of compound 19.
<html><A name="NMR19H"/></html>

<html><A name="NMR19F"/></html>

1H NMR and 19F NMR of compound 21.
<html><A name="NMR21H"/></html>

<html><A name="NMR21F"/></html>

Peptide lock
Spectra
(S)-2-((tert-butoxycarbonyl)amino)-5-methoxy-5-oxopentanoic acid
1H NMR

13C NMR

<html></div></div></html> <html> <head> <style>
- indexing {
/* float: left; position: center; */ background-color: #222; border-top: 2px solid #d13f31; color: #006e9c; margin: 0px; padding: 0px 0px 10px 0px; width: 100%; text-align: center; }
.footer-section { padding: 10px; display: table-cell; text-align: left; }
.footer-section-title { font-size: 20px; }
- footer-contents {
color: #006e9c; display: inline-table; }
.footer-section A { color: #006e9c; text-decoration: none; }
.footer-section A:HOVER { color: #00aeef; }
.footer-section ul { list-style-type: square; }
- sitemapTitle {
margin-top: 20px; font-size: 24px; }
- editFooter {
float: right; margin-top: -28px; margin-right: 5px; }
- editFooter A {
color: #006e9c; text-decoration: none; }
.cf:before,.cf:after { content: " "; /* 1 */ display: table; /* 2 */ }
.cf:after { clear: both; }
- bodyContent a[href^="mailto:"], .link-mailto {
background: url() no-repeat scroll right center transparent; padding-right: 0px; color: #006e9c;
}
</style> </head> <body> <div id="indexing"> <div id="sitemap"> <p id="sitemapTitle">SITEMAP | BIOMOD 2013 NANO CREATORS | Aarhus University</p> <div id="footer-contents"> <div class="footer-section"> <p class="footer-section-title">INTRODUCTION</p> <ul> <li><a href="/wiki/Biomod/2013/Aarhus">Home, abstract, animation and video</a></li> <li><a href="/wiki/Biomod/2013/Aarhus/Introduction">Introduction</a></li </ul> </div> <div class="footer-section"> <p class="footer-section-title">RESULTS AND DISCUSSION</p> <ul> <li><a href="/wiki/Biomod/2013/Aarhus/Results_And_Discussion/Origami">Origami</a></li> <li><a href="/wiki/Biomod/2013/Aarhus/Results_And_Discussion/Peptide_lock">Peptide lock</a></li> <li><a href="/wiki/Biomod/2013/Aarhus/Results_And_Discussion/Chemical_Modification">Chemical modification</a></li> <li><a href="/wiki/Biomod/2013/Aarhus/Results_And_Discussion/sisiRNA">sisiRNA</a></li> <li><a href="/wiki/Biomod/2013/Aarhus/Results_And_Discussion/System_In_Action">System in action</a></li> </ul> </div> <div class="footer-section"> <p class="footer-section-title">MATERIALS AND METHODS</p> <ul> <li><a href="/wiki/Biomod/2013/Aarhus/Materials_And_Methods/Origami">Origami</a></li> <li><a href="/wiki/Biomod/2013/Aarhus/Materials_And_Methods/Peptide_lock">Peptide lock</a></li> <li><a href="/wiki/Biomod/2013/Aarhus/Materials_And_Methods/Chemical_Modification">Chemical modification</a></li> <li><a href="/wiki/Biomod/2013/Aarhus/Materials_And_Methods/sisiRNA">sisiRNA</a></li> <li><a href="/wiki/Biomod/2013/Aarhus/Materials_And_Methods/System_In_Action">System in action</a></li> <li><a href="/wiki/Biomod/2013/Aarhus/Materials_And_Methods/Methods">Methods</a></li> </ul> </div> <div class="footer-section"> <p class="footer-section-title">SUPPLEMENTARY</p> <ul> <li><a href="/wiki/Biomod/2013/Aarhus/Supplementary/Team_And_Acknowledgments">Team and acknowledgments</a></li> <li><a href="/wiki/Biomod/2013/Aarhus/Supplementary/Optimizations">Optimizations</a></li> <li><a href="/wiki/Biomod/2013/Aarhus/Supplementary/Supplementary_Data">Supplementary data</a></li>
<li><a
href="/wiki/Biomod/2013/Aarhus/Supplementary/Supplementary_Informations">Supplementary informations</a> <li><a href="/wiki/Biomod/2013/Aarhus/Supplementary/References">References</a></li> </ul> </div> </div> <div> <p id="copyright">Copyright (C) 2013 | BIOMOD Team Nano Creators @ Aarhus University | Programming by: <a href="mailto:pvskaarup@gmail.com?Subject=BIOMOD 2013:">Peter Vium Skaarup</a>.</p> </div> </div>
<!-- Sponsers --> <div> <img alt="Sigma - Aldrich" src="http://openwetware.org/images/3/39/Sigmaaldrich-logo%28transparant%29.png" width="300px" height="154px"> <img alt="VWR International" src="http://openwetware.org/images/2/28/Vwr_logo.png" width="300px" height="61px"> <img alt="Promega" src="http://openwetware.org/images/7/72/Promega.png" width="175px" height="105px" style="padding-right: 5px; padding-left: 5px;"> <img alt="kem-en-tec" src="http://openwetware.org/images/3/3a/Kementec.png" width="130px" height="129px"> <img alt="Centre For Dna Nanotechnology" src="http://openwetware.org/images/4/4f/CDNA_logo.png" width="420px" height="90px"> <img alt="Dansk Tennis Fond" src="http://openwetware.org/images/9/9a/Dansk_tennis.png" width="250px" height="53px"> </div> <div id="editFooter"> [<a href="http://openwetware.org/index.php?title=Template:Biomod/2013/Aarhus/Nano_Creators/footer&action=edit">edit sitemap</a>] </div> </div> </body> </html>



