BioSysBio:abstracts/2007/Gokhan Karakulah
- Add or delete the sections that you require.
miRNA EXPRESSION AND SEQUENCE DATABASE
Author(s): Kaya K. D.1, Karakulah G.2, Konu O.1, Yakicier C.1
Affiliations: 1Department of Molecular Biology and Genetics, Bilkent University, Ankara,Turkey.
2Department of Medical Informatics, Health Sciences Institute, Dokuz Eylul
University, Izmir, Turkey.
Contact:email: kkaya@bilkent.edu.tr, gokhan.karakulah@deu.edu.tr
Keywords: 'miRNA' 'dinucleotide' 'expression' 'systems biology'
Background/Introduction
MicroRNAs are small ribonucleic acids, which bind to 3'UTR regions of mRNAs by base complementation. They play crucial roles in the regulation of embryonic development, and in many cellular events such as proliferation, apoptosis, and differentiation. Hence, understanding the mechanisms by which they are regulated is a key attractant for research intentions involving microRNAs. Accordingly we have investigated the significance of the bias in dinucleotide motif distribution of miRNAs by using a randomization approach. The correlation of sequence characteristics with miRNA microarray expression values was also statistically assessed. Our results indicated that observed miRNA frequencies of multiple dinucleotide motifs significantly deviated from expected and their relative abundancies are close to genomic values. Moreover, variation in the level of miRNA expression could partly be explained by the presence or absence of a CpG motif. Our study is the development of a web interfaced database, which integrates the species-specific mature miRNA sequence information and the associated public microarray data and outputs tabular and graphical summaries after statistical calculations (through R). The database also summarizes GO annotation network data of the targets of miRNAs that are in question. Our database is still in progress and it will have more user friendly capabilities in near future http://139.179.97.62/~koray.
Results
This database put miRNA targets and public expression data together with an oportunity to select miRNAs according to their dinucleotide properties. It gives also a chance to follow the gaps about their roles. We tested our data base with different miRNAs of specific expression pattern. We know from the literature that mir-15a and mir-16 are expressed in bone marrow and spleen more specifically. Again from the literature, they are deleted in Chronic Lymphocytic Leukemia. One of the annotated targets of them is BCL-2, a well known antiapoptotic protein. Our database catches the term “anti-apoptotic” as a significant term.
First we selected a human expression data, and then selected the two miRNAs.

After submission we get the expression pattern.


And we selected biological process category and clicked the get go data button. And the result is:

As it can be followed from the table, anti-apoptotic and regulation of apoptosis terms gave highly low p values of a hypergeometric test. Another significant term is humoral immune response, however when we searched it in pubmed, we got:

From our previous studies we know that the expression of CpG containing miRNAs are less expressed in all tissues except brain. And we know from the literature mamalian genome is scarce in CpG dinucleotide. We wanted to see whether this scarcity is reflected to function or not. Here we present the differences. The initial expectation was that we could have significant terms for CpG bearing miRNAs. What we have found is the opposite what we expected.
Non-CpG
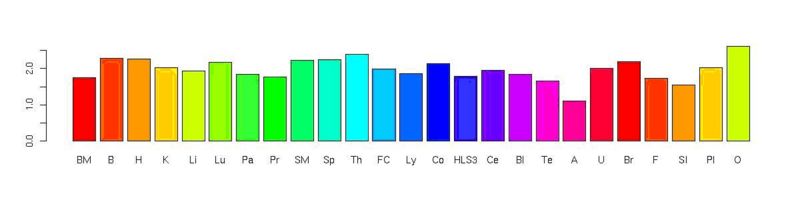

CpG


In order to see that if there is a bias in the miRNA set used in the Baskerville data, we selected all miRNAs in the data set. And what we got is :

Conclusion
These results may mean that non-CpG miRNAs target the genes that cover almost all most frequent biologic function ontologies among miRNA targets, because it is the way of having low p values for the frequent terms among targets in geometric distribution. On the other hand, CpG containing miRNAs have more versatile targets other than the targets with common ontologies. So, this may still show existance of attributable regulatory roles to CpG containing miRNAs. Further classification of CpG miRNAs, i.e., according to tissues where they are expressed prominently, may revael the possible biologic functions specific to CpG miRNAs. However there may be also a bias due to the incomplete data.
The both significant terms as the result of biologic function ontology analysis of mir-15a and mir-16, antiapoptotis and humoral immune response, come from the same gene, BCL-2, and there is not a direct relation however between and immune response. Though we consider the cancer as a multistep progression disease, the deficiency in miR-15a or 16 may be the cause of some immune diseases.
In similar ways, many research focuses can be created through utilization of our database. Due to the time constraints, however, we can not demonstrate more examples altough possible. We are improving our database still, and it will be integrated with more system biology tools in the future. As being improved, it will help understanding miRNAs in the whole system context more.
References
[1] Karlin, S. and J. Mrazek, 1997, Compositional differences within and between eukaryotic genomes, Proc Natl Acad Sci U S A 94, 10227.
[2] Bartel, D.P., 2004, MicroRNAs: genomics, biogenesis, mechanism, and function, Cell 116, 281.
[3] Baskerville, S. and D.P. Bartel, 2005, Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes, Rna 11, 241.
[4] Thomson, J.M., J. Parker, C.M. Perou and S.M. Hammond, 2004, A custom microarray platform for analysis of microRNA gene expression, Nat Methods 1, 47.
[5] Carney D. A., Wierda W. G., 2005 May, Genetics and molecular biology of chronic lymphocytic leukemia, Curr Treat Options Oncol. (3):215-25. Links
[6] Shahi P. et al., 2006 Jan, Argonaute--a database for gene regulation by mammalian microRNAs, Nucleic Acids Res. 34(Database issue): D115-8