BioMicroCenter:Nextera: Difference between revisions
No edit summary |
No edit summary |
||
| (26 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{{BioMicroCenter}} | |||
[[Image: | [[Image:Nextera Illumina fig1.gif|right]]Nextera DNA sample preparation, from Epicentre technologies (now a subsidiary of Illumina), uses a modified TN5 transposase to simultaneously fragment intact genomic DNA and tag it with Illumina adapters. A limited number of PCR steps are required to generate complete Illumina libraries.<br><br> | ||
Nextera | |||
The Nextera | The BioMicro Center offers three flavors of Nextera preps, depending on input and throughput requirements. Original nextera is the most expensive, requiring 50ng of input material. Products can be sized by [[BioMicroCenter:Pippin|pippin]] and are suitable for all applications. NexteraXT uses less input material (1ng) and, thus, less enzyme making it less expensive, but only crude size fractionation using single-pass SPRI selection can be done. Finally, in November 2014, we introduced an automated version of the Nextera XT kit that is done on our [[BioMicroCenter:Tecan|Tecan EVOs]]. These are done in batches of 16 with some failure rate tolarated (~10%). <BR><BR> | ||
{|style="border:1px solid purple" | |||
! | |||
! Nextera | |||
! NexteraXT | |||
! Automated | |||
|- | |||
|'''Input''' | |||
|50ng | |||
|1ng | |||
|1ng | |||
|- | |||
|'''Sizing''' | |||
|Pippin or SPRI | |||
|SPRI | |||
|SPRI | |||
|- | |||
|'''#samples''' | |||
|1 | |||
|1 | |||
|16/batch | |||
|- | |||
|} | |||
[[Image:CNV_data_example.png| | Nextera-prepared samples provided a similar quality of sequencing data compared with samples prepped in parallel on the SPRI-Works system. DNA from the same Colobacter sample was either sonicated for SPRI-prep or provided as intact DNA for Nextera prep. The samples were multiplexed and sequenced together on an Illumina GAII 40bp single-end lane. The total genomic coverage for both Nextera and SPRI-te samples was exactly the same at 97.8% coverage of Colobacter’s GC-rich genome, although complexity was greater in the sonicated samples. | ||
[[Image:Nextera_2.png|550px]]<br><br> | |||
[[Image:Nextera_3.png|left]] [[Image:Nextera_4.png|300px|right]]The Nextera transposase does exhibit a mild GC-insertion bias, shown by the increase in percent of the first few bases. This, however, did not affect the sequencing coverage. The Nextera-prepared sample has very consistent genome-wide coverage, while the SPRI-prepared samples have a more variable coverage density. | |||
<br><br><br><br><br><br><br><br>We have used Nextera DNA sample preparation for multiple samples and have seen high-quality data for all the samples. Below is an example of a CNV study prepared with Nextera DNA preparation:<br> | |||
[[Image:CNV_data_example.png|1000px]] | |||
Latest revision as of 17:35, 13 October 2014
| HOME -- | SEQUENCING -- | LIBRARY PREP -- | HIGH-THROUGHPUT -- | COMPUTING -- | OTHER TECHNOLOGY |
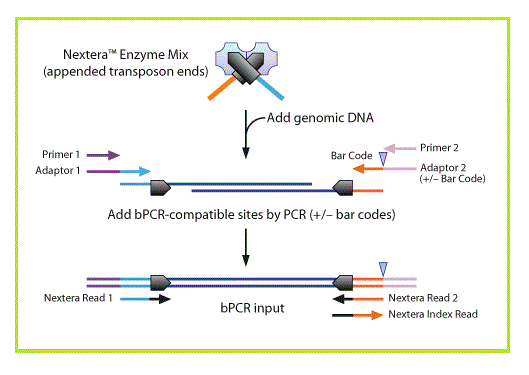
Nextera DNA sample preparation, from Epicentre technologies (now a subsidiary of Illumina), uses a modified TN5 transposase to simultaneously fragment intact genomic DNA and tag it with Illumina adapters. A limited number of PCR steps are required to generate complete Illumina libraries.
The BioMicro Center offers three flavors of Nextera preps, depending on input and throughput requirements. Original nextera is the most expensive, requiring 50ng of input material. Products can be sized by pippin and are suitable for all applications. NexteraXT uses less input material (1ng) and, thus, less enzyme making it less expensive, but only crude size fractionation using single-pass SPRI selection can be done. Finally, in November 2014, we introduced an automated version of the Nextera XT kit that is done on our Tecan EVOs. These are done in batches of 16 with some failure rate tolarated (~10%).
| Nextera | NexteraXT | Automated | |
|---|---|---|---|
| Input | 50ng | 1ng | 1ng |
| Sizing | Pippin or SPRI | SPRI | SPRI |
| #samples | 1 | 1 | 16/batch |
Nextera-prepared samples provided a similar quality of sequencing data compared with samples prepped in parallel on the SPRI-Works system. DNA from the same Colobacter sample was either sonicated for SPRI-prep or provided as intact DNA for Nextera prep. The samples were multiplexed and sequenced together on an Illumina GAII 40bp single-end lane. The total genomic coverage for both Nextera and SPRI-te samples was exactly the same at 97.8% coverage of Colobacter’s GC-rich genome, although complexity was greater in the sonicated samples.

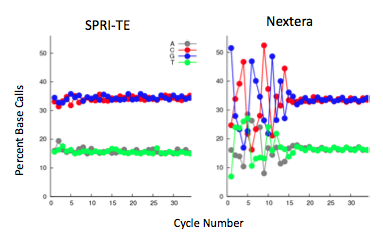

The Nextera transposase does exhibit a mild GC-insertion bias, shown by the increase in percent of the first few bases. This, however, did not affect the sequencing coverage. The Nextera-prepared sample has very consistent genome-wide coverage, while the SPRI-prepared samples have a more variable coverage density.
We have used Nextera DNA sample preparation for multiple samples and have seen high-quality data for all the samples. Below is an example of a CNV study prepared with Nextera DNA preparation:

