BioMicroCenter:Nextera: Difference between revisions
No edit summary |
No edit summary |
||
| (16 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
[[Image: | [[Image:Nextera Illumina fig1.gif|right]]Nextera DNA sample preparation, from Epicentre technologies, uses a modified transposase to simultaneously fragment intact genomic DNA and tag it with Illumina adapters. A limited number of PCR steps are required to generate complete Illumina libraries.<br><br> | ||
Nextera-prepared samples provided a similar quality of sequencing data compared with samples prepped in parallel on the SPRI-Works system. DNA from the same Colobacter sample was either sonicated for SPRI-prep or provided as intact DNA for Nextera prep. The samples were multiplexed and sequenced together on an Illumina GAII 40bp single-end lane. The total genomic coverage for both Nextera and SPRI-te samples was exactly the same at 97.8% coverage of Colobacter’s GC-rich genome, although complexity was greater in the sonicated samples. | Nextera-prepared samples provided a similar quality of sequencing data compared with samples prepped in parallel on the SPRI-Works system. DNA from the same Colobacter sample was either sonicated for SPRI-prep or provided as intact DNA for Nextera prep. The samples were multiplexed and sequenced together on an Illumina GAII 40bp single-end lane. The total genomic coverage for both Nextera and SPRI-te samples was exactly the same at 97.8% coverage of Colobacter’s GC-rich genome, although complexity was greater in the sonicated samples. | ||
[[Image:Nextera_2.png | [[Image:Nextera_2.png|550px]]<br><br> | ||
The Nextera transposase does exhibit a mild GC-insertion bias, shown by the increase in percent of the first few bases. This, however, did not affect the sequencing coverage. The Nextera-prepared sample has very consistent genome-wide coverage, while the SPRI-prepared samples have a more variable coverage density. | [[Image:Nextera_3.png|left]] [[Image:Nextera_4.png|300px|right]]The Nextera transposase does exhibit a mild GC-insertion bias, shown by the increase in percent of the first few bases. This, however, did not affect the sequencing coverage. The Nextera-prepared sample has very consistent genome-wide coverage, while the SPRI-prepared samples have a more variable coverage density. | ||
<br><br><br><br><br><br><br><br>We have used Nextera DNA sample preparation for multiple samples and have seen high-quality data for all the samples. Below is an example of a CNV study prepared with Nextera DNA preparation:<br> | |||
[[Image:CNV_data_example.png|1000px]] | |||
[[Image:CNV_data_example.png| | |||
Revision as of 09:32, 29 September 2011
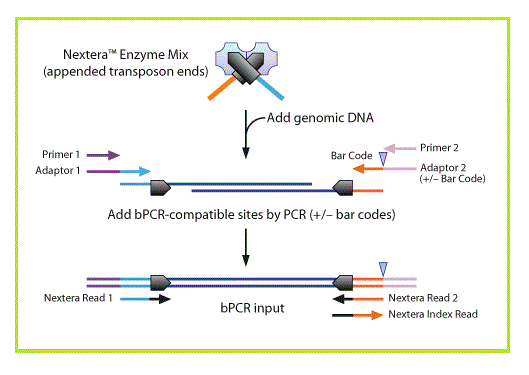
Nextera DNA sample preparation, from Epicentre technologies, uses a modified transposase to simultaneously fragment intact genomic DNA and tag it with Illumina adapters. A limited number of PCR steps are required to generate complete Illumina libraries.
Nextera-prepared samples provided a similar quality of sequencing data compared with samples prepped in parallel on the SPRI-Works system. DNA from the same Colobacter sample was either sonicated for SPRI-prep or provided as intact DNA for Nextera prep. The samples were multiplexed and sequenced together on an Illumina GAII 40bp single-end lane. The total genomic coverage for both Nextera and SPRI-te samples was exactly the same at 97.8% coverage of Colobacter’s GC-rich genome, although complexity was greater in the sonicated samples.

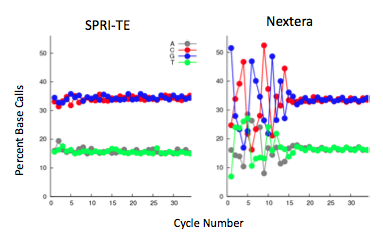

The Nextera transposase does exhibit a mild GC-insertion bias, shown by the increase in percent of the first few bases. This, however, did not affect the sequencing coverage. The Nextera-prepared sample has very consistent genome-wide coverage, while the SPRI-prepared samples have a more variable coverage density.
We have used Nextera DNA sample preparation for multiple samples and have seen high-quality data for all the samples. Below is an example of a CNV study prepared with Nextera DNA preparation:
