BioMicroCenter:DNA LIB: Difference between revisions
No edit summary |
|||
| Line 14: | Line 14: | ||
| 150-350, 250-550 or 100+ | | 150-350, 250-550 or 100+ | ||
|- | |- | ||
| Neoprep | | [[BioMicroCenter:DNA_LIB#NEOPREP|Neoprep]] | ||
| LM-PCR | | LM-PCR | ||
| 1ng - 100ng | | 1ng - 100ng | ||
| 250+ only | | 250+ only | ||
|- | |- | ||
| Nextera / NexteraXT | | [[BioMicroCenter:DNA_LIB#NEXTERA_.2F_NEXTERAXT|Nextera / NexteraXT]] | ||
| Fragmentation with Tn5 transposase | | Fragmentation with Tn5 transposase | ||
| 50ng / 1ng | | 50ng / 1ng | ||
Revision as of 11:07, 9 August 2016
| HOME -- | SEQUENCING -- | LIBRARY PREP -- | HIGH-THROUGHPUT -- | COMPUTING -- | OTHER TECHNOLOGY |
The BioMicro Center offers a broad variety of methodologies for preparing DNA libraries for sequencing. As will all of our standard prep methods, pricing includes quality control prior and following prep, along with at least one re-prep for samples that fail. For large batches of samples or 16S/amplicon, please visit our High-Throughput DNA library preparation page.
| Method | Summary | Input | Insert Range |
|---|---|---|---|
| SPRIworks | LM-PCR | 100pg - 100ng | 150-350, 250-550 or 100+ |
| Neoprep | LM-PCR | 1ng - 100ng | 250+ only |
| Nextera / NexteraXT | Fragmentation with Tn5 transposase | 50ng / 1ng | adjustable minimum by SPRI cleanup |
SPRI WORK

Fragmented samples submitted to the BioMicro Center are processed using the Beckman Coulter SPRIworks. This system preforms the sample cleanup, a overhang, ligation and preliminary size selection needed for creating Illumina libraries. The system accepts all amounts of insert and has been extensively tested.
The SPRI-te Nucleic Acid Extractor is a fully-automated library preparation system for Illumina sequencing and supports all Illumina applications except for smallRNA-Seq. It performs all of the traditional Illumina protocol steps listed on the Illumina Library Preparation page with the exception of PCR enrichment. A total of 10 libraries may be prepared simultaneously in 5 hours.
BASICS
|
The SPRI-Works system consists of three main components: the SPRI-TE Nucleic Acid Extractor, a method card that contains the liquid handling program, and a cartridge containing all of the reagents required to prepare a single library. The platform utilizes AMPureXP DNA-binding magnetic beads to purify fragmented DNA and perform size selection, eliminating the need for column purification and gel-based size selection. After the adapter ligation and size selection steps are complete, samples are ready for PCR enrichment and clean-up to complete the library prep process. For more information on the SPRI-Works system please visit www.spriworks.com. |
 |
Size Selection
Library Complexity
Effect of Concentration
Sample Submission
We suggest that you submit as much volume as possible since the first step of the protocol brings all sample volumes up to 100uL in water. For optimal yield, libraries should be fragmented so that the average fragment size is approximately 60nt shorter than the chosen size range to allow for the addition of partial adapters.
All samples submitted for BMC Fragmented DNA sample prep (SPRI-only) must be submitted with user supplied adapter mix. Adapter mixes should be pre-diluted and should have enough volume for 10uL of adapter per sample. For recommended dilutions for various sample concentrations, please see the chart below:

Please note that SPRI-only samples will need to be re-submitted separately if you wish to sequence with us later on.
NEOPREP DNA
NEXTERA / NEXTERAXT

Nextera DNA sample preparation, from Epicentre technologies (now a subsidiary of Illumina), is our preferred method for preparing Illumina libraries from intact DNA. Nextera uses a modified TN5 transposase to simultaneously fragment intact genomic DNA and tag it with Illumina adapters. A limited number of PCR steps are required to generate complete Illumina libraries. This very simple method makes this preparation very popular for automation and in vivo methods such as ATACseq.
The BioMicro Center offers three flavors of Nextera preps, depending on input and throughput requirements. Original Nextera is the most expensive, requiring 50ng of input material but produces the most complex libraries. Products can be sized by pippin and are suitable for all applications. NexteraXT uses less input material (1ng) and, thus, less enzyme making it less expensive, but only crude size fractionation using single-pass SPRI selection can be done. Finally, in November 2014, we introduced an automated version of the Nextera XT kit that is done on our Tecan EVOs. For more information about high-throuhgput Nextera, please look at the High-Throuhgput DNA preparation page. The BMC does not offer ATACseq as that works directly with the intact cells.
Nextera works well with amplicons as well as gDNA. However, because two hits are required per molecule to create productive libraries, the ratio of reagent to fragment must be altered significantly to produce good libraries and large inserts may not be possible.
| Nextera | NexteraXT | |
|---|---|---|
| Input | 50ng | 1ng |
| Sizing | Pippin or SPRI | SPRI |
Example Data

Nextera-prepared samples provided a similar quality of sequencing data compared with samples prepped in parallel on the SPRI-Works system. DNA from the same Colobacter sample was either sonicated for SPRI-prep or provided as intact DNA for Nextera prep. The samples were multiplexed and sequenced together on an Illumina GAII 40bp single-end lane. The total genomic coverage for both Nextera and SPRI-te samples was exactly the same at 97.8% coverage of Colobacter’s GC-rich genome, although complexity was greater in the sonicated samples.
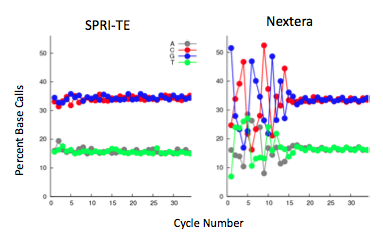
The Nextera transposase does exhibit a mild GC-insertion bias, shown by the increase in percent of the first few bases. Generally, this has a minimal impact on sequencing coverage, though some variation will occur. Below is an example of a CNV study prepared with Nextera DNA preparation:






