Banta:BetaRoll: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
{{Template:Banta_Lab}} | {{Template:Banta_Lab}} | ||
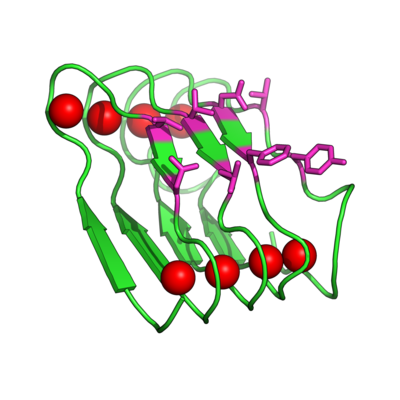
[[Image:BetaRoll.jpg|thumb|400px|right|Crystal structure of a folded Beta Roll domain where the red spheres indicate the bound Ca+2 ions (Dooley, Kim, Lu, Tu, and Banta, 2012 Biomacromolecules).]] | [[Image:BetaRoll.jpg|thumb|400px|right|Crystal structure of a folded Beta Roll domain where the red spheres indicate the bound Ca+2 ions and the purple resides are randomized for molecular recognition (Dooley, Kim, Lu, Tu, and Banta, 2012 Biomacromolecules).]] | ||
'''Evolving the Beta Roll Domain for Regulated Molecular Recognition''' | '''Evolving the Beta Roll Domain for Regulated Molecular Recognition''' | ||
Revision as of 11:33, 8 October 2012

Banta Lab
Protein and Metabolic Engineering
| Home | Lab Members | Publications | Research Interests | Courses | Pictures | Positions Available |
Evolving the Beta Roll Domain for Regulated Molecular Recognition
Molecular recognition is ubiquitous in nature. Frequently antibodies are used in technology applications where biomolecular recognition is to be employed, but antibodies have several limitations in these applications, including difficulty in easily removing bound targets. This becomes especially important in the development of biosensors using electrochemical-based signal transduction schemes.
Instead of trying to engineer allosteric control into a molecular recognition molecule, we have started with intrinsically disordered scaffold, the beta roll domain, and we are working to evolve this allosterically regulated scaffold for biomolecular recognition. The naturally existing beta roll subdomain motif consists of tandem repeats of the sequence GGXGXDXUX, where U is an aliphatic amino acid and X is any amino acid. In the presence of calcium, the disordered peptide reversibly transitions to a beta roll spiral structure of two parallel beta sheet faces, where each beta strand has two solvent exposed variable residues.
We have characterized a native beta roll subdomain with various end-capping groups in order to identify a minimal calcium-responsive beta roll unit. We believe that the beta roll faces are suitable binding surfaces and that calcium-induced structure formation can be used as a mechanism to control the formation of the engineered biomolecular recognition interface. The reversibility of the calcium binding suggests that the engineered biomolecular recognition will likewise be reversibly controllable. We have randomized one face of this minimal beta roll unit and we are using directed evolution to identify beta roll peptides with biomolecular recognition capabilities.
Related Publications
- Shur O, Wu J, Cropek DM, and Banta S. Monitoring the conformational changes of an intrinsically disordered peptide using a quartz crystal microbalance. Protein Sci. 2011 May;20(5):925-30. DOI:10.1002/pro.625 |
- Blenner MA, Shur O, Szilvay GR, Cropek DM, and Banta S. Calcium-induced folding of a beta roll motif requires C-terminal entropic stabilization. J Mol Biol. 2010 Jul 9;400(2):244-56. DOI:10.1016/j.jmb.2010.04.056 |
- Szilvay GR, Blenner MA, Shur O, Cropek DM, and Banta S. A FRET-based method for probing the conformational behavior of an intrinsically disordered repeat domain from Bordetella pertussis adenylate cyclase. Biochemistry. 2009 Dec 1;48(47):11273-82. DOI:10.1021/bi901447j |
- Chockalingam K, Blenner M, and Banta S. Design and application of stimulus-responsive peptide systems. Protein Eng Des Sel. 2007 Apr;20(4):155-61. DOI:10.1093/protein/gzm008 |
- Banta S, Megeed Z, Casali M, Rege K, and Yarmush ML. Engineering protein and peptide building blocks for nanotechnology. J Nanosci Nanotechnol. 2007 Feb;7(2):387-401. DOI:10.1166/jnn.2007.153 |