BME103:T930 Group 3: Difference between revisions
Rohan Kumar (talk | contribs) |
Rohan Kumar (talk | contribs) |
||
| Line 123: | Line 123: | ||
<!--- Place two small Image J data images here. One showing the result of Water and the other showing the result of Calf Thymus DNA ---> | <!--- Place two small Image J data images here. One showing the result of Water and the other showing the result of Calf Thymus DNA ---> | ||
[[Image:BME-3_Data.png]]<br> | |||
<!--- Enter the values from your group's Data Analyzer table below. E6, F6, etc. are the excel cells from which you should copy your data. ---> | <!--- Enter the values from your group's Data Analyzer table below. E6, F6, etc. are the excel cells from which you should copy your data. ---> | ||
Revision as of 21:58, 14 November 2012
| Home People Lab Write-Up 1 Lab Write-Up 2 Lab Write-Up 3 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
OUR TEAM
LAB 1 WRITE-UPInitial Machine TestingThe Original Design
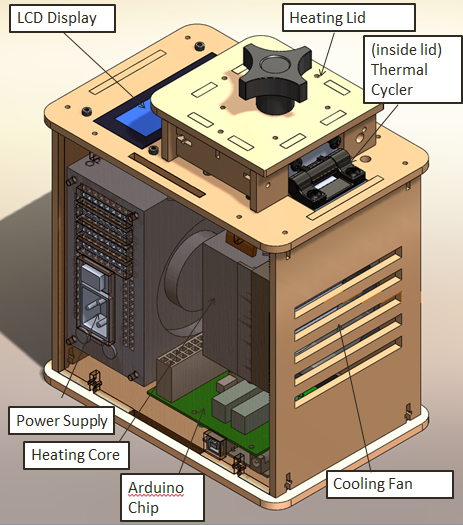
When we unplugged the lcd display wire(part 3) from the Arudnio chip(part 6), the screen turned off. Everything on the PCR was working fine expect there was no output on the display. When we unplugged the white wire that connects Arudino chip(part 6) to thermal cycler (part 2), the reading from the screen dropped to -40 degrees Celsius. We disconnected the wire multiple times and each time the screen displayed -40 degrees Celsius.
We ran a test run on 10/25/2012. For this test we placed some empty PCR tubes into the machine and ran a simple test program on the Open PCR software. After the simple test was over we noticed that the display screen on the Open PCR lid matched very closely with what was displayed on our computer screen. The agreement between our computer screen and our PCR display meant that our diagnostic test was a success.
ProtocolsPolymerase Chain Reaction Polymerase Chain Reaction Procedure: 2.)We labeled 8 empty PCR tubes. For the first sample we labeled the 3 DNA samples 1A, 1B and 1C. For the second sample we labeled the tubes 2A, 2B and 2C. For the positive and negative controls, we labeled the tubes + and - respectively. 3.)Using one pipette per sample, to avoid contamination, we transferred the PCR reaction mix we were given to the PCR tubes. 4.)We then placed the samples in the PCR machine 5.)We set our PCR program to three stages. Stage one: 1 cycle, 95 degree Celsius for 3 minutes. Stage 2: 35 cycles, 95 degrees Celsius for 30 seconds, 57 degrees Celsius for 30 seconds, 72 degrees Celsius. Stage three: 72 degrees Celsius for 3 minutes and then hold at 4 degree Celsius.
Sample one ID 43891: 48 Male GO Taq DNA Mix
Fluorimeter assembly Procedure:
Research and DevelopmentSpecific Cancer Marker Detection - The Underlying Technology (Add a write-up of the information discussed in Week 3's class) (BONUS points: Use a program like Powerpoint, Word, Illustrator, Microsoft Paint, etc. to illustrate how primers bind to the cancer DNA template, and how Taq polymerases amplify the DNA. Screen-captures from the OpenPCR tutorial might be useful. Be sure to credit the source if you borrow images.)
Results
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||