BME100 s2015:Group1 9amL5: Difference between revisions
| (57 intermediate revisions by 5 users not shown) | |||
| Line 15: | Line 15: | ||
|- valign="top" | |- valign="top" | ||
| [[Image:shawnstriker.jpg|100px|thumb|Name: Shawn Striker]] | | [[Image:shawnstriker.jpg|100px|thumb|Name: Shawn Striker]] | ||
| [[Image: | | [[Image:JonathanMoreno.jpg|100px|thumb|Name: Jonathan Moreno]] | ||
| [[Image: | | [[Image:GraceKim.jpg|100px|thumb|Name: Grace Kim]] | ||
| [[Image: | | [[Image:Triston Hudson.jpg|100px|thumb|Name: Triston Hudson]] | ||
| [[Image: | | [[Image:RyanJMeyer.jpg|100px|thumb|Name: Ryan Meyer]] | ||
| [[Image:BME103student.jpg|100px|thumb|Name: student]] | | [[Image:BME103student.jpg|100px|thumb|Name: student]] | ||
|} | |} | ||
| Line 37: | Line 37: | ||
** Saturation: N/A | ** Saturation: N/A | ||
** Contrast: N/A | ** Contrast: N/A | ||
Settings that are N/A: Camera is sensitive enough to get a reasonable calibration with DNA concentration | Settings that are N/A: <br> | ||
Camera is sensitive enough to get a reasonable calibration with DNA concentration <br> | |||
Camera was focused when necessary | |||
'''Calibration'''<br> | '''Calibration'''<br> | ||
<!-- INSTRUCTIONS: In the space below, briefly describe how to set up your camera in front of the fluorimeter. Add a PHOTO of this set-up for bonus points. --> | <!-- INSTRUCTIONS: In the space below, briefly describe how to set up your camera in front of the fluorimeter. Add a PHOTO of this set-up for bonus points. --> | ||
#Turn on the camera on the smartphone and calibrate according to the settings above | #Turn on the camera on the smartphone and calibrate according to the settings above | ||
#Place the smartphone in the cradle | #Place the smartphone in the cradle | ||
#Place the cradle at a right angle to the slide | #Place the cradle at a right angle to the slide | ||
# | #Adjust the height of the fluorimeter using the plastic trays in order to take a picture of the drop sideways | ||
#Adjust the distance between the cradle with the smartphone and the slide so that the cradle is at least 4 cm away from the slide | |||
#Adjust the distance between the cradle with the smartphone and the slide | #Record the distance between the cradle and the slide with a ruler | ||
<!-- INSTRUCTIONS: Type the distance between your phone cradle and the drop after the equal sign. --> | <!-- INSTRUCTIONS: Type the distance between your phone cradle and the drop after the equal sign. --> | ||
* Distance between the smart phone cradle and drop = 8cm | * Distance between the smart phone cradle and drop = 8cm | ||
'''Solutions Used for Calibration''' | '''Solutions Used for Calibration''' | ||
[[image:chart54.png]] | |||
*Water at pH = 8 | |||
*SYBR green I dye solution | |||
*DNA solution | |||
'''Placing Samples onto the Fluorimeter''' | '''Placing Samples onto the Fluorimeter''' | ||
# | #Use the pipettor to place a drop of the SYBR green I dye solution (80 microliters) in the middle of the first two rows of the slide | ||
# | #Use the pipettor to add a drop of the water blank solution (80 microliters) | ||
# | #Adjust the slide so that the drop focuses the blue LED light to the middle of the black fiber optic fitting on the other side of the drop | ||
# | #Cover the fluorimeter and camera with the light box so that all stray light is removed | ||
#Take three pictures and make sure the drop is focused | |||
#Remove the box | |||
#Use the pipettor to remove the drop from the slide (160 microliters) | |||
#Move the slide to the next position | |||
#Repeat steps 1-8 for the patients and the DNA solution | |||
<br> | <br> | ||
| Line 81: | Line 85: | ||
<!-- INSTRUCTIONS: (1) Show ONE image where you drew a circle around the droplet with the freehand tool for any sample with *no* DNA. (2) Show ONE image where you drew a circle around the droplet with the freehand tool for a sample *with* DNA (positive signal). -If you include more than two images, you will not receive any additional credit. --> | <!-- INSTRUCTIONS: (1) Show ONE image where you drew a circle around the droplet with the freehand tool for any sample with *no* DNA. (2) Show ONE image where you drew a circle around the droplet with the freehand tool for a sample *with* DNA (positive signal). -If you include more than two images, you will not receive any additional credit. --> | ||
[[image: | Positive Sample | ||
[[image:positive3.png|400px]] | |||
Negative Sample | |||
[[image:negtive3.png|400px]] | |||
<br> | |||
<br> | |||
Patient 1-1 | |||
[[image:one1.png|400px|]] | |||
Patient 1-2 | |||
[[image:one2.png|400px|]] | |||
Patient 1-3 | |||
[[image:one3.png|400px|]] | |||
Patient 2-1 | |||
[[image:two1.png|400px|]] | |||
Patient 2-2 | |||
[[image:two2.png|400px|]] | |||
Patient 2-3 | |||
[[image:two3.png|400px|]] | |||
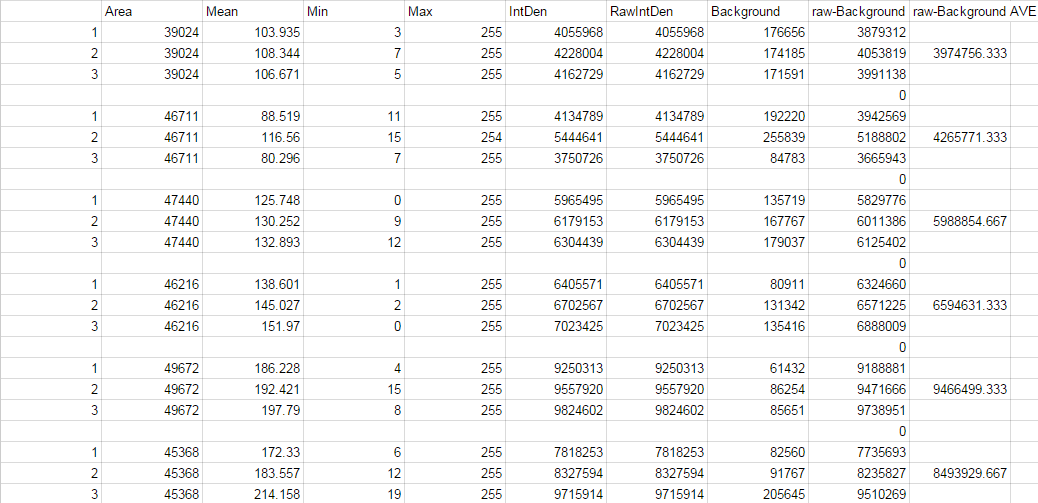
'''Image J Values for All Calibrator Samples''' | '''Image J Values for All Calibrator Samples''' | ||
| Line 90: | Line 118: | ||
[[image: | [[image:dsfa.png]] | ||
'''Calibration curve'''<br> | '''Calibration curve'''<br> | ||
<!-- INSTRUCTIONS: Place an image of your Excel plot with a line of best fit here. --> | <!-- INSTRUCTIONS: Place an image of your Excel plot with a line of best fit here. --> | ||
[[Image:cal333.png]] | |||
'''PCR Results Summary''' | '''PCR Results Summary''' | ||
<!-- INSTRUCTIONS: You completed 8 PCR reactions and used the SYBR Green I staining and imaging technique to measure the amount of amplified DNA in each PCR reaction. You used a standard curve (based on known concentrations of calf thymus DNA) to convert INTDEN values into DNA concentration. Your positive control and negative control samples should be used as '''threshold''' values for determining whether an unknown (patient) sample is truly positive or negative. Replace the underscore with your claculated initial concentration values.--> | <!-- INSTRUCTIONS: You completed 8 PCR reactions and used the SYBR Green I staining and imaging technique to measure the amount of amplified DNA in each PCR reaction. You used a standard curve (based on known concentrations of calf thymus DNA) to convert INTDEN values into DNA concentration. Your positive control and negative control samples should be used as '''threshold''' values for determining whether an unknown (patient) sample is truly positive or negative. Replace the underscore with your claculated initial concentration values.--> | ||
* Our positive control PCR result was | * Our positive control PCR result was 0.05596862783 μg/mL | ||
* Our negative control PCR result was | * Our negative control PCR result was 0.02701367497 μg/mL | ||
[[Image:doobaadoo.png]] | |||
<u>Observed results</u> | <u>Observed results</u> | ||
<!-- INSTRUCTIONS: Replace the underscore with each patient ID. After the colon, write both a qualitative (what the images looked like) and a quantitative description (μg/mL) of what you observed --> | <!-- INSTRUCTIONS: Replace the underscore with each patient ID. After the colon, write both a qualitative (what the images looked like) and a quantitative description (μg/mL) of what you observed --> | ||
* Patient | * Patient 11316 : There is a notable amount of the SYBR green I dye solution in the drop. | ||
* Patient | 0.01599345468 μg/mL | ||
* Patient 25353 : Compared to patient 11316, the amount of SYBR green I dye solution in the drop is insignificant. There is still a significant amount present, however, patient 11316 shows more of the dye. | |||
0.02401010622 μg/mL | |||
<u>Conclusions</u> | <u>Conclusions</u> | ||
<!-- INSTRUCTIONS: Compare each patient's results to the positive control value and the negative control value. Draw a final conclusion for each patient (positive or negative) and explain why you made that conclusion. --> | <!-- INSTRUCTIONS: Compare each patient's results to the positive control value and the negative control value. Draw a final conclusion for each patient (positive or negative) and explain why you made that conclusion. --> | ||
* Patient | * Patient 11316 : It was concluded that the patient did not contract the disease because the PCR concentration is close to the the negative control PCR result. | ||
* Patient | * Patient 25353 : It was concluded that the patient did not contract the disease because the amount of dye present in the drop is less than the amount of dye present in the drop for patient 11316. Both the PCR concentrations in the drops are extremely close to the negative control value amount, however patient 25353 shows a lesser amount (closer to the negative control) | ||
| Line 119: | Line 152: | ||
'''Background: About the Disease SNP''' | '''Background: About the Disease SNP''' | ||
<!-- INSTRUCTIONS: This content is from PCR Lab D. Write a summary, at least five sentences long, about the disease SNP in your own words. --> | <!-- INSTRUCTIONS: This content is from PCR Lab D. Write a summary, at least five sentences long, about the disease SNP in your own words. --> | ||
SNPs, or single nucleotide polymorphisms, are an affliction on one's DNA. They are the most common source of genetic variation found among HomoSapiens, or the human population. <br> | |||
Essentially a SNP takes one nucleotide and changes it into a different one. For example, if the nucleotide is supposed to be an A, an SNP will change the A into a T. <br> | |||
By changing nucleotides, variations within humans arise, or the differences in humans' phenotypes (appearances). | |||
<br> | |||
<br> | |||
Question answers from the lab manual: | |||
<br> | |||
*Nucleotide- “A nucleotide is one of the structural components, or building blocks, of DNA and RNA. | |||
A nucleotide consists of a base (one of four chemicals: adenine, thymine, guanine, and cytosine) plus a molecule of sugar and one of phosphoric acid.” | |||
Taken from: http://www.ncbi.nlm.nih.gov/Class/MLACourse/Original8Hour/Genetics/nucleotide.html | |||
<br> | |||
*Polymorphism-“Single nucleotide polymorphisms, frequently called SNPs (pronounced “snips”), are the most common type of genetic variation among people. | |||
Each SNP represents a difference in a single DNA building block, called a nucleotide.” | |||
Taken from: http://ghr.nlm.nih.gov/handbook/genomicresearch/snp | |||
<br> | |||
rs268 | |||
*What species is the variation found in? – Homo Sapiens | |||
<br> | |||
*What chromosome is the variation located on? - 8:19956018 | |||
<br> | |||
*What is listed as the Clinical significance of this SNP? – Pathogenic | |||
<br> | |||
*Which gene(s) is this SNP associated with? – LPL | |||
<br> | |||
*Click the PubMed link to view summaries of research associated with the SNP. What disease is linked to this SNP? – Metabolic syndrome susceptibility | |||
<br> | |||
*What does LPL stand for? – Lipoprotein lipase | |||
<br> | |||
*What is the function of LPL? To find out, click the LPL link. Look for “Gene ontology” in the right hand list and click it. Write the first three unique terms you see... | |||
1) apolipoprotein binding | |||
2) heparin binding | |||
3) chylomic ron remodeling | |||
<br> | |||
*What is an allele? – “An allele is an alternative form of a gene(one member of a pair) that is located at a specific position on a specific chromosome. These DNA codings determine distinct traits that can be passed on from parents to offspring throughsexual reproduction.” Taken from: http://biology.about.com/od/geneticsglossary/g/alleles.htm | |||
<br> | |||
*The disease-associated allele contains what sequence? – A G T | |||
<br> | |||
*The Numerical position of the SNP is -19956018 | |||
<br> | |||
*Non-disease forward primer (20 nt): A A T C T G G G C T A T G A G A T C A A | |||
<br> | |||
*The numerical position exactly 200 bases to the right of the disease SNP is: 19956219 | |||
<br> | |||
*Non-disease reverse primer (20 nt): A A T G C A A C C C C C T A T C A A C A G | |||
<br> | |||
*Disease forward primer (20 nt): A A T C T G G G C T A T G A G A T C A G | |||
<br> | |||
*Disease reverse primer: same as non-disease reverse primer | |||
'''Primer Design and Testing''' | '''Primer Design and Testing''' | ||
<!-- INSTRUCTIONS: Write a short summary of the results of your primer test. Underneath your summary, include a screen capture of the results web page. You may crop the image so that it only includes the relevant information. --> | <!-- INSTRUCTIONS: Write a short summary of the results of your primer test. Underneath your summary, include a screen capture of the results web page. You may crop the image so that it only includes the relevant information. --> | ||
[[Image:screenshot1.jpeg]] | |||
[[Image:screenshot2.jpeg]] | |||
Latest revision as of 09:53, 8 April 2015
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |||||||
|
OUR TEAM
LAB 5 WRITE-UPProcedureSmart Phone Camera Settings
Settings that are N/A:
Solutions Used for Calibration
Placing Samples onto the Fluorimeter
Data AnalysisRepresentative Images of Negative and Positive Samples
Image J Values for All Calibrator Samples Calibration curve PCR Results Summary
Observed results
0.01599345468 μg/mL
0.02401010622 μg/mL Conclusions
SNP Information & Primer DesignBackground: About the Disease SNP
SNPs, or single nucleotide polymorphisms, are an affliction on one's DNA. They are the most common source of genetic variation found among HomoSapiens, or the human population.
A nucleotide consists of a base (one of four chemicals: adenine, thymine, guanine, and cytosine) plus a molecule of sugar and one of phosphoric acid.” Taken from: http://www.ncbi.nlm.nih.gov/Class/MLACourse/Original8Hour/Genetics/nucleotide.html
Each SNP represents a difference in a single DNA building block, called a nucleotide.” Taken from: http://ghr.nlm.nih.gov/handbook/genomicresearch/snp
1) apolipoprotein binding 2) heparin binding 3) chylomic ron remodeling
| |||||||