BME100 s2015:Group16 12pmL5: Difference between revisions
No edit summary |
|||
| (27 intermediate revisions by 3 users not shown) | |||
| Line 40: | Line 40: | ||
'''Calibration'''<br> | '''Calibration'''<br> | ||
<!-- INSTRUCTIONS: In the space below, briefly describe how to set up your camera in front of the fluorimeter. Add a PHOTO of this set-up for bonus points. --> | <!-- INSTRUCTIONS: In the space below, briefly describe how to set up your camera in front of the fluorimeter. Add a PHOTO of this set-up for bonus points. --> | ||
[[Image:g16calibration.png]] | |||
<!-- INSTRUCTIONS: Type the distance between your phone cradle and the drop after the equal sign. --> | <!-- INSTRUCTIONS: Type the distance between your phone cradle and the drop after the equal sign. --> | ||
| Line 68: | Line 70: | ||
'''Placing Samples onto the Fluorimeter''' | '''Placing Samples onto the Fluorimeter''' | ||
# '' | # ''Turn on Blue LED Excitation light'' | ||
# '' | # ''Insert a slide into the Fluorimeter'' | ||
# '' | # ''Place camera on the craddle and as close to as possible without getting a blurry picture'' | ||
# '' | # ''Adjust height of fluorimeter so that picture from camera takes a picture of the drop directly from the side'' | ||
# ''Place an 80 microliter drop of SYBR Green I on the slide so that it looks like a beach ball'' | |||
# ''Add an 80 microliter drop of the DNA solution to the already placed solution'' | |||
# ''Align the slide so that the blue LED is focused on the drop'' | |||
# ''Take the a picture with the timer set and lower the lid of the lightbox'' | |||
# ''Take a total of 3 pictures then remove the solution from the slide'' | |||
# ''Move the slide to the next position and repeat the process with the different solutions'' | |||
<br> | <br> | ||
| Line 81: | Line 89: | ||
<!-- INSTRUCTIONS: (1) Show ONE image where you drew a circle around the droplet with the freehand tool for any sample with *no* DNA. (2) Show ONE image where you drew a circle around the droplet with the freehand tool for a sample *with* DNA (positive signal). -If you include more than two images, you will not receive any additional credit. --> | <!-- INSTRUCTIONS: (1) Show ONE image where you drew a circle around the droplet with the freehand tool for any sample with *no* DNA. (2) Show ONE image where you drew a circle around the droplet with the freehand tool for a sample *with* DNA (positive signal). -If you include more than two images, you will not receive any additional credit. --> | ||
Positve: | |||
[[Image:positvejpg.jpg]] | |||
Negative: | |||
[[Image:negativejpg.jpg]] | |||
| Line 87: | Line 103: | ||
{| | |||
| align="center" style="background:#f0f0f0;"|'''Final DNA concentration in SYBR Green I solution (μg/mL)''' | |||
| align="center" style="background:#f0f0f0;"|'''Area''' | |||
| align="center" style="background:#f0f0f0;"|'''Mean Pixel Value''' | |||
| align="center" style="background:#f0f0f0;"|'''RawIntDen of the drop''' | |||
| align="center" style="background:#f0f0f0;"|'''RawIntDen of the background''' | |||
| align="center" style="background:#f0f0f0;"|'''RawIntDen drop - background''' | |||
|- | |||
| 2.5||139380||108.933||15183035||37084||15145951 | |||
|- | |||
| 2.5||142636||109.163||15570617||37393||15533224 | |||
|- | |||
| 2.5||147752||107.308||15854933||49037||15805896 | |||
|- | |||
| 1||106708||83.381||8897448||17939||8879509 | |||
|- | |||
| 1||115956||80.25||9305435||21998||9283437 | |||
|- | |||
| 1||113112||90.715||10260974||34066||10226908 | |||
|- | |||
| 0.5||135592||62.432||8465278||64454||8400824 | |||
|- | |||
| 0.5||118410||58.929||6977741||64902||6912839 | |||
|- | |||
| 0.5||115368||58.331||6729582||46971||6682611 | |||
|- | |||
| 0.25||139968||40.608||5683780||59072||5624708 | |||
|- | |||
| 0.25||127236||40.933||5208100||54709||5153391 | |||
|- | |||
| 0.25||133808||42.526||5690375||51402||5638973 | |||
|- | |||
| 0.125||131884||38.479||5074747||49047||5025700 | |||
|- | |||
| 0.125||124424||34.43||4283904||43969||4239935 | |||
|- | |||
| 0.125||132332||37.836||5006942||54115||4952827 | |||
|- | |||
| 0||137400||21.711||2983023||52412||2930611 | |||
|- | |||
| 0||124400||18.681||2323902||45159||2278743 | |||
|- | |||
| 0||114528||19.95||2284888||39148||2245740 | |||
|} | |||
{| | |||
| align="center" style="background:#f0f0f0;"|'''Final DNA concentration in SYBR Green I solution (μg/mL)''' | |||
| align="center" style="background:#f0f0f0;"|'''RawIntDen drop - background''' | |||
| align="center" style="background:#f0f0f0;"|'''''' | |||
| align="center" style="background:#f0f0f0;"|'''''' | |||
| align="center" style="background:#f0f0f0;"|'''''' | |||
| align="center" style="background:#f0f0f0;"|'''Standard Deviation''' | |||
|- | |||
| ||1||2||3||mean|| | |||
|- | |||
| 2.5||15145951||15533224||15805896||46485071||331626.7478 | |||
|- | |||
| 1||8879509||9283437||10226908||28389854||691469.3802 | |||
|- | |||
| 0.5||8400824||6912839||6682611||21996274||932680.7426 | |||
|- | |||
| 0.25||5624708||5153391||5638973||16417072||276325.015 | |||
|- | |||
| 0.125||5025700||4239935||4952827||14218462||434156.6754 | |||
|- | |||
| 0||2930611||2278743||2245740||7455094||386235.9758 | |||
|} | |||
'''Calibration curve'''<br> | '''Calibration curve'''<br> | ||
<!-- INSTRUCTIONS: Place an image of your Excel plot with a line of best fit here. --> | <!-- INSTRUCTIONS: Place an image of your Excel plot with a line of best fit here. --> | ||
[[Image:g16calcurve.png]] | |||
'''PCR Results Summary''' | '''PCR Results Summary''' | ||
<!-- INSTRUCTIONS: You completed 8 PCR reactions and used the SYBR Green I staining and imaging technique to measure the amount of amplified DNA in each PCR reaction. You used a standard curve (based on known concentrations of calf thymus DNA) to convert INTDEN values into DNA concentration. Your positive control and negative control samples should be used as '''threshold''' values for determining whether an unknown (patient) sample is truly positive or negative. Replace the underscore with your claculated initial concentration values.--> | <!-- INSTRUCTIONS: You completed 8 PCR reactions and used the SYBR Green I staining and imaging technique to measure the amount of amplified DNA in each PCR reaction. You used a standard curve (based on known concentrations of calf thymus DNA) to convert INTDEN values into DNA concentration. Your positive control and negative control samples should be used as '''threshold''' values for determining whether an unknown (patient) sample is truly positive or negative. Replace the underscore with your claculated initial concentration values.--> | ||
* Our positive control PCR result was | * Our positive control PCR result was .11603 μg/mL | ||
* Our negative control PCR result was | * Our negative control PCR result was .0575 μg/mL | ||
<u>Observed results</u> | <u>Observed results</u> | ||
<!-- INSTRUCTIONS: Replace the underscore with each patient ID. After the colon, write both a qualitative (what the images looked like) and a quantitative description (μg/mL) of what you observed --> | <!-- INSTRUCTIONS: Replace the underscore with each patient ID. After the colon, write both a qualitative (what the images looked like) and a quantitative description (μg/mL) of what you observed --> | ||
* Patient | * Patient 47360 : Average initial concentration of .114425, the bubble observed was clearer than the positive control, however much darker than the negative | ||
* Patient | * Patient 31303 : Average initial concentration of .15928, the bubble was much darker the negative control and the patient 1, much closer to the color of the positive control. | ||
<u>Conclusions</u> | <u>Conclusions</u> | ||
<!-- INSTRUCTIONS: Compare each patient's results to the positive control value and the negative control value. Draw a final conclusion for each patient (positive or negative) and explain why you made that conclusion. --> | <!-- INSTRUCTIONS: Compare each patient's results to the positive control value and the negative control value. Draw a final conclusion for each patient (positive or negative) and explain why you made that conclusion. --> | ||
* Patient | * Patient POS : 2 out of three of the runs ran closer in similarity to the positive control | ||
* Patient | * Patient POS : All three of the runs were closer to the positive test | ||
| Line 117: | Line 204: | ||
'''Background: About the Disease SNP''' | '''Background: About the Disease SNP''' | ||
<!-- INSTRUCTIONS: This content is from PCR Lab D. Write a summary, at least five sentences long, about the disease SNP in your own words. --> | <!-- INSTRUCTIONS: This content is from PCR Lab D. Write a summary, at least five sentences long, about the disease SNP in your own words. --> | ||
An SNP is a single nucleotide polymorphism(SNP). An SNP occurs when a nucleotide pair, the monomers that create DNA, is altered changing the gene it is contained in creating a polymorphism, a genetic variation. These SNPs occur naturally in biological genomes and through natural selection these mutations are controlled. When an SNP occurs it creates a new allele, a variation in the gene, in a population that if not detrimental will be passed on to the next generation. It is usual for there to be only two different alleles for a specific gene which is the case for the SNP being studied. The disease SNP being studied is rs268. This SNP occurs in Homo Sapiens on the eighth chromosome. The clinical significance of this SNP is that it is pathogenic and it is associated with the LPL gene, the lipoprotein lipase. This gene is involved with processes such as apolipoprotein binding, chylomicron remodeling, and anchoring components of the membrane. This disease is responsible for metabolic syndrome that increase blood pressure, cholesterol levels, excess fat, and blood sugar levels which increases a person risk for stroke, diabetes, and heart disease. The normal allele of this gene is AAT but when the disease is present the mutation creates an allele of AGT. | |||
'''Primer Design and Testing''' | '''Primer Design and Testing''' | ||
<!-- INSTRUCTIONS: Write a short summary of the results of your primer test. Underneath your summary, include a screen capture of the results web page. You may crop the image so that it only includes the relevant information. --> | <!-- INSTRUCTIONS: Write a short summary of the results of your primer test. Underneath your summary, include a screen capture of the results web page. You may crop the image so that it only includes the relevant information. --> | ||
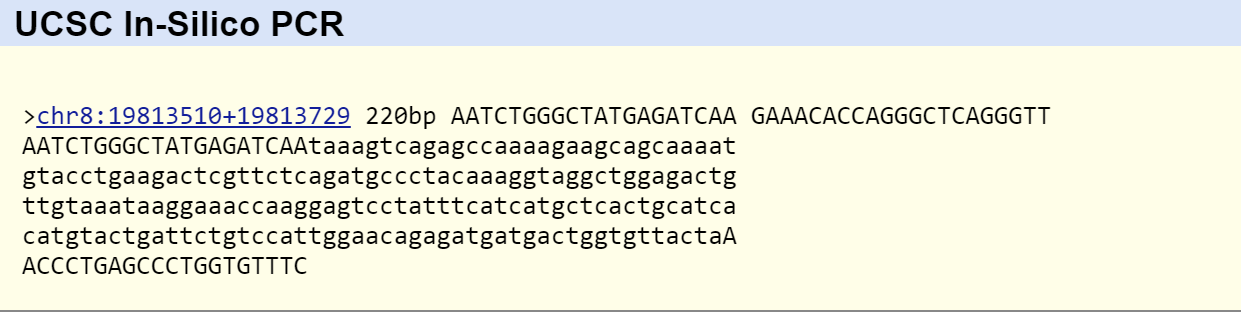
The primers worked as expected producing one positive test and one negative test. In the positive test the diseased primer was shown to work. In the negative test the non-disease primer was shown to work. The primers worked by bonding to the DNA and allowing replication to occur if they matched properly. By getting replicated DNA it was confirmed that the primers worked because they were attached. The primers were different at two different points in the forward direction. The non-disease primer would bond allowing replication if the allele was negative for the disease because it ended in an 'A' nucleotide which the gene has if it does not have the disease. The disease primer bonded and allowed replication to occur if the diseased allele was because it ended in a 'G' which the gene would have in at that location if the disease was present. The two nucleotide primers can be seen below. | |||
'''Non-Disease Primer''' | |||
[[Image:Primer Valid.jpg]] | |||
'''Disease Primer''' | |||
[[Image:Primer Invalid.jpg]] | |||
<!-- Do not edit below this line --> | <!-- Do not edit below this line --> | ||
|} | |} | ||
Latest revision as of 23:59, 7 April 2015
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 5 WRITE-UPProcedureSmart Phone Camera Settings
Data AnalysisRepresentative Images of Negative and Positive Samples Positve:
Observed results
Conclusions
SNP Information & Primer DesignBackground: About the Disease SNP An SNP is a single nucleotide polymorphism(SNP). An SNP occurs when a nucleotide pair, the monomers that create DNA, is altered changing the gene it is contained in creating a polymorphism, a genetic variation. These SNPs occur naturally in biological genomes and through natural selection these mutations are controlled. When an SNP occurs it creates a new allele, a variation in the gene, in a population that if not detrimental will be passed on to the next generation. It is usual for there to be only two different alleles for a specific gene which is the case for the SNP being studied. The disease SNP being studied is rs268. This SNP occurs in Homo Sapiens on the eighth chromosome. The clinical significance of this SNP is that it is pathogenic and it is associated with the LPL gene, the lipoprotein lipase. This gene is involved with processes such as apolipoprotein binding, chylomicron remodeling, and anchoring components of the membrane. This disease is responsible for metabolic syndrome that increase blood pressure, cholesterol levels, excess fat, and blood sugar levels which increases a person risk for stroke, diabetes, and heart disease. The normal allele of this gene is AAT but when the disease is present the mutation creates an allele of AGT.
The primers worked as expected producing one positive test and one negative test. In the positive test the diseased primer was shown to work. In the negative test the non-disease primer was shown to work. The primers worked by bonding to the DNA and allowing replication to occur if they matched properly. By getting replicated DNA it was confirmed that the primers worked because they were attached. The primers were different at two different points in the forward direction. The non-disease primer would bond allowing replication if the allele was negative for the disease because it ended in an 'A' nucleotide which the gene has if it does not have the disease. The disease primer bonded and allowed replication to occur if the diseased allele was because it ended in a 'G' which the gene would have in at that location if the disease was present. The two nucleotide primers can be seen below. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||