BME100 s2014:W Group5 L6: Difference between revisions
| Line 37: | Line 37: | ||
'''Our Design'''<br> | '''Our Design'''<br> | ||
<p>[[Image:[NewLid1.png]]</p> | |||
<p>[[Image:NewLid2.png]]</p> | |||
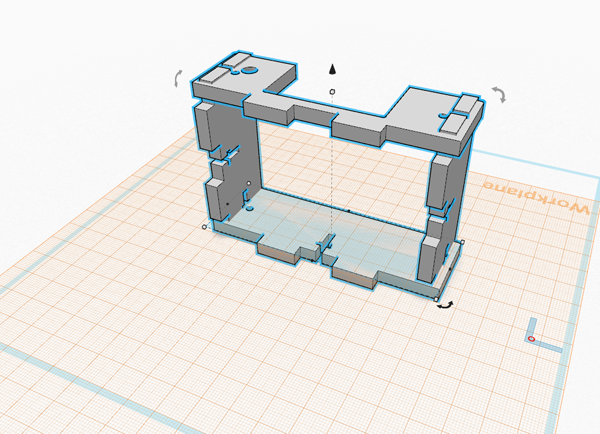
<p>We modified the design by manipulating the material used to build the OpenPCR machine rather than the shape or functionality of any given part. </p> | <p>We modified the design by manipulating the material used to build the OpenPCR machine rather than the shape or functionality of any given part. </p> | ||
Revision as of 11:40, 16 April 2014
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |
|
OUR COMPANY[Instructions: add the name of your team's company and/or product here]
LAB 6 WRITE-UPComputer-Aided DesignTinkerCAD TinkerCAD is a web-based computer-aided drafting programme that allows the users to design three-dimensional objects. These objects can then be manipulated in size, shape and colour before being sent to a 3D printer and output as a real, physical object.
[[Image:[NewLid1.png]] We modified the design by manipulating the material used to build the OpenPCR machine rather than the shape or functionality of any given part. One of the more frustrating parts of working with the OpenPCR was the difficulty we had in finding the optimal tight- or snug-ness between the heated lid and the samples underneath it. In changing the material used to build the side walls of the lid to something more transparent (yet still retaining the conductive properties of the metal), the user will not need to estimate so much when tightening the lid over the samples (or having it pop open after tightening the lid too far)
Feature 1: Disease SNP-Specific Primers[Instructions: This information will come from the exercises you did in PCR Lab B.] Background on the cancer-associated mutation [Instructions: Use the answers from questions 3 - 7 to compose, in your own words, a paragraph about rs237025]
Primer design
How the primers work: [Instructions: explain what makes the primers disease-sequence specific. In other words, explain why the primers will amplify DNA that contains the cancer-associated SNP, and will not exponentially amplify DNA that has the non-disease allele.]
Feature 2: Consumables KitOne of the major issues we had with the consumables was the issue of SYBR green bleaching. Repeatedly covering and uncovering the entire set of samples got tedious after 20+ samples and merely putting foil over the top of the samples does not prevent reflected light from coming in through the bottom. To fix this issue, we plan to re-package the consumables in the kit with a black, light-proof tube (or tubes) for the SYBR green dye. This will allow the user to leave the tubes uncovered without running the risk of ruining the test.
Feature 3: Hardware - PCR Machine & Fluorimeter[Instructions: Summarize how you will include the PCR machine and fluorimeter in your system. You may add a schematic image. An image is OPTIONAL and will not get bonus points, but it will make your report look really awesome and easy to score.] [Instructions: IF your group has decided to redesign the PCR machine and/or Fluorimeter to address any major weakness(es), explain how in an additional paragraph.]
Bonus Opportunity: What Bayesian Stats Imply About The BME100 Diagnostic Approach[Instructions: This section is OPTIONAL, and will get bonus points if answered thoroughly and correctly. Here is a chance to flex some intellectual muscle. In your own words, discuss what the results for calculations 3 and 4 imply about the reliability of PCR for predicting the disease. Please do NOT type the actual numerical values here. Just refer to them as being "less than one" or "very small." The instructors will ask you to submit your actual calculations via a Blackboard quiz. We are doing so for the sake of academic integrity and to curb any temptation to cheat.]
| |