BISC220/S14: Mod 1 Lab 2
Enzyme Structure/Function: Background Information
The purification of an enzyme is often required in order to study its physical characteristics and the reaction it catalyzes. Purification is essentially a concentrating process in which the concentration of the target enzyme is increased while other proteins are eliminated. The enrichment process also eliminates metabolites and other cellular constituents that may influence the enzyme structure and/or the reaction. Background information on protein purification can be found below and also in your textbook.
During our first lab series, we will be partially purifying and characterizing the enzyme β-galactosidase from Escherichia coli, strain BL21(pET-14b). Although we will not be concerned with regulation of gene expression, it is of interest that experimentation on the production of β-galactosidase by E. coli ultimately led Francois Jacob and Jacques Monod to propose the 'operon' model for the regulation of gene expression. If you are interested in learning more about transcription regulation and early work on the genetic analysis of the lac-operon in E. coli, your textbook is a good resource.
To study protein structure and function, we have chosen the enzyme β-galactosidase for a variety of reasons. First, it is a convenient enzyme for learning basic techniques in enzymology because it is readily available in large quantities from microbial cells and is relatively stable. Next, the gene has been fully sequenced and cloned. The crystal structure of the protein and hence its 3-D configuration has been solved. Finally, the specific activity assay generally used to assess enzyme function is reliable and convenient (Jacobson et al., 1994; Ring, 1990; Wallenfels and Weil, 1972).
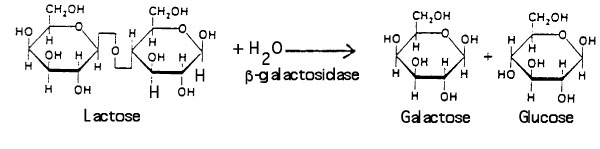
Βeta-galactosidase allows E. coli to grow in a culture medium containing the sugar lactose as the sole source of energy. The enzyme catalyzes the hydrolysis of lactose into glucose and galactose:
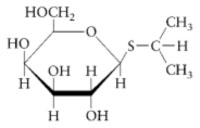
The purification of β-galactosidase is facilitated when E. coli produce large quantities of the enzyme. Growth on lactose as a carbon source is not a sufficient means of insuring abundant production of the enzyme since the expression of β-galactosidase is controlled. β-galactosidase production remains partially repressed even when cells are grown in the presence of the substrate lactose. It is possible, however, to transform genetically engineered strains of E. coli with a plasmid that expresses high levels of β-galactosidase. The plasmid fuses a strong promoter upstream of the β-galactosidase gene, lacZ. The gene promoter (and the gene’s over expression) is regulated by isopropyl-β-D-thiogalactoside (IPTG), the structure of which is shown below.
In addition to allowing the production of large quantities of β-galactosidase, the lacZ gene on the plasmid, is modified so that the primary sequence of the enzyme is extended on the amino terminus with six histidines. This 6xHis tag can be exploited to purify the protein using metal chelate affinity chromatography (Kagedal, 1998), which we will perform in Lab 2.
Our goals for Lab 1 are:
- Calibrate our micropipets
- Induce E. coli BL21(pET-14b) cells to produce large quantities of 6xHis tagged β-galactosidase
- Learn how to model proteins and analyze their sequences, using the computer programs RasMol and Clustal Omega.
References:
Jacobson RH, Zhang XJ, DuBose RF, Matthews BW (1994) Three-dimensional structure of β-galactosidase from E. coli. Nature. 369:761-6.
Karp G (2005) Cell and Molecular Biology Concepts and Experiments, 4th edition, John Wiley & Sons, Inc.,Hoboken.
Ring M, Huber RE (1990) Multiple replacements establish the importance of tyrosine-503 in beta-galactosidase (Escherichia coli). Arch Biochem Biophys. 283:342-50.
Wallenfels K, Weil R (1972) “β-galactosidase” In The Enzymes (Boyer PD, ed), Vol VII, 3rd ed, Academic Press, New York, pp 617-663.
Tutorials
RasMol Tutorial
ClustalW Tutorial
Protocols
THE INDUCTION OF β-GALACTOSIDASE
You will be provided with approximately 200 ml of E. coli BL21 (pET-14b) in log phase growth in Luria broth (LB) media. You will induce the cells of this genetically engineered strain to increase production of 6xHis-β-galactosidase by adding IPTG to the cells and incubating them at 37°C for one hour. You will collect the cells by centrifugation and freeze the cell pellet until the next lab period.
- Remove a flask containing the E. coli BL21(pET-14) cells from the 37°C shaking incubator and bring it to your lab bench.
- Using a sterile 1 ml pipette and aseptic technique, remove 1.0 ml of the culture and add it to a microfuge tube labeled "pre-IPTG cells" on tape of your team color. Please also include the initials of your group. Centrifuge this small aliquot of cells for 2 minutes in a microcentrifuge. Discard the supernatant into the waste container next to the centrifuge and give the labeled dry pellet to your instructor to freeze at -70°C.
- Using a sterile 5.0 ml pipette and aseptic technique, add 2.0 ml of 0.1M IPTG to the flask of growing E. coli cells. This addition will yield a final concentration of approximately 1.0 mM IPTG in the culture flask.
- Return the flask of E. coli cells containing IPTG to the incubator and allow the cells to incubate with shaking for 60 minutes. During this time IPTG will induce production of large amounts of β-galactosidase.
- Calibrate your micropipets while your protein is synthesized by our genetically engineered strain of E. coli. Your instructor will demonstrate how to use the micropipets. Then follow the calibration protocol on the next page.
- After the incubation period, remove the flask of bacterial cells from the incubator. Remove 1.0 ml of the culture to a microfuge tube labeled "+ IPTG cells". Make sure the label includes the initials of your lab group, team color, & lab day. Pellet the cells as in step 2 above. Give the labeled tube containing the dry pellet to your instructor for freezing at -70°C.
- Pour the post-induction cells into an empty centrifuge bottle with a screw-cap provided by your lab instructor; secure the cap. Do not overfill the centrifuge bottle. If you have extra culture, it should be left in the flask to be autoclaved before disposal. Please do not throw any bacterial cells down the drain!
- Centrifuge the cells for 15 minutes at 8,000 RPM (~10,400x g) in a big Sorval centrifuge using the GSA rotor. Be sure that your bottle is balanced with another group’s before starting the centrifuge. Use sterile LB media for equalizing the weights.
- Carefully pour the supernatant back into the flask. Remove with a Pasteur pipet any media that remains. Label the bottle containing your cell pellet with the name of the cells (E. coli BL21(pET-14) + IPTG) on a piece of tape in your team color and include your initials and lab day; give your instructor the bottle to freeze at -70°C until the next lab period.
For a Word™ format protocol: Media:PROTOCOL FOR THE INDUCTION OF βgal.doc
Protocol for Micropipette Calibration
- To calibrate your P1000, P 200, and P 20 micropipets, label 6 microfuge tubes (1-6) and weigh them. Record the weights in the table below.
- Following the table below, pipet the specified volumes into the pre-weighed microfuge tubes prepared above and then re-weigh them. Record all weights.
- Calculate the weight of the water in grams. 1000 microliters of water should weigh 1 gram at room temperature.
- If the water in any tube weighs more or less than 1 gram, ask your instructor for help. If your calibration is significantly off after several repeated attempts, your pipet (or your technique!) may need adjustment.
| Tube # | Tube Pre-Weight | Vol. in μL using P20 | Vol. in μL using P200 | Vol. in μL using P1000 | Weight of Tube + Water in grams | Weight of Water in grams |
|---|---|---|---|---|---|---|
For a Word™ format protocol: Media:Protocol for Micropipet Calibration.doc
