BISC209/F13: Lab8: Difference between revisions
Tucker Crum (talk | contribs) |
Tucker Crum (talk | contribs) |
||
| Line 127: | Line 127: | ||
'''Formic Acid Extraction Procedure: Tube Extraction (TE)''' <BR> | '''Formic Acid Extraction Procedure: Tube Extraction (TE)''' <BR> | ||
Materials Needed :<BR> | |||
<UL><LI>Eppendorf 1.5 μL microfuge tubes | <UL><LI>Eppendorf 1.5 μL microfuge tubes | ||
<LI>Eppendorf pipettes/tips | <LI>Eppendorf pipettes/tips | ||
| Line 141: | Line 140: | ||
<LI>Formic Acid, 70% HPLC/MS Grade | <LI>Formic Acid, 70% HPLC/MS Grade | ||
<LI>Acetonitrile, 100 % HPLC/MS Grade | <LI>Acetonitrile, 100 % HPLC/MS Grade | ||
<LI>Fresh Microorganism: overnight growth should be used for routine microorganism identification; slow-growing bacteria may need to cultivate for several days before testing; do not use organisms that have been stored at 4°C or lower as this has a negative impact on quality of spectra and reproducibility; storing plates at room temperature for several days is acceptable; different media types and growth temperatures have little effect on results.</LI></UL></UL | <LI>Fresh Microorganism: overnight growth should be used for routine microorganism identification; slow-growing bacteria may need to cultivate for several days before testing; do not use organisms that have been stored at 4°C or lower as this has a negative impact on quality of spectra and reproducibility; storing plates at room temperature for several days is acceptable; different media types and growth temperatures have little effect on results.</LI></UL></UL><BR> | ||
# Label a microfuge tube for each of your isolates on the side of the tube using a graphite pencil. Put the code on the top of the tube also using a Sharpie. Use a unique isolate code, for example A2-1 for soil sample and isolate number. <BR> | # Label a microfuge tube for each of your isolates on the side of the tube using a graphite pencil. Put the code on the top of the tube also using a Sharpie. Use a unique isolate code, for example A2-1 for soil sample and isolate number. <BR> | ||
# Add 300 μL of HPLC water to each microfuge tube <BR> | # Add 300 μL of HPLC water to each microfuge tube <BR> | ||
Revision as of 12:02, 24 October 2013
Lab 8: Con't Tests on Cultured Isolates
Finding Evidence for Co-operation and Competition Among Cultured Members of a Soil Community
Con't Testing for Antibiotic Production
Week 2
TO BE PROVIDED:
3 fresh cultures of Eschericia coli (Gram negative), Staphylococcus epidermidis (Gram positive) and Micrococcus luteus (Gram positive) grown in nutrient broth to a McFarland 5 standard turbidity.
PROTOCOL
-
Use a sterile swab and aseptically apply a line of inoculation of each of the provided broth cultures of : E. coli, Micrococcus, and S. epidermidis.
as shown below. Draw a line perpendicular to the antibiotic producer's (Streptomyces) inoculation. Be careful not to touch the antibiotic producer's growth. Using the same swabs, inoculate a new NA plate (one plate for all three cultures)by making a line across the plate for E. coli, Micrococcus and S. epidermidis. Incubate this plate along with your test plate. It will serve as a control to make sure that lack of growth is due to antibiotic sensitivity and not to no living cells in the inoculum.

- Incubate for another week.
Complete the Interaction Assays
If you haven't been provided with a digital image, please take a photo of each of your interaction plates. Make sure that the photos are sharp enough to evaluate later. Download them to your dropbox in Sakai. Email them to yourself and to each of your partners.
A sample completed assay is shown below.
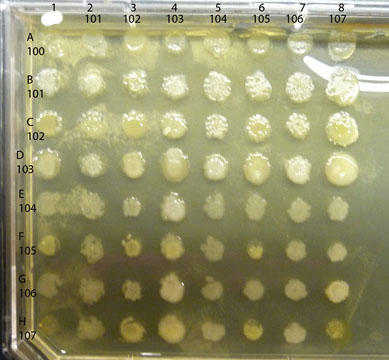
Observe the colonies on your plate, comparing any differences in the appearance of the colony growth of each isolate, alone vs mixed. Look first at each control culture: the inoculum in each of the diagonal spots is a pure culture control as are the spots in the column on the far left. Compare each "spot" where two isolates are mixed to the control spots where each isolate is growing alone. Is either isolate growing better in combination than alone? If so, you have found a mutualistic (beneficial) interaction. For example, in the assay shown above, isolate #102 appears to have a positive effect on the growth of #101, classified as mutualism. Are there combinations that show a reduction in the amount of growth of either isolate compared to the growth of the control areas? If so, you have found an antagonistic (negative) interaction. An example in the assay plate shown above is between isolates #105 and #102. Number 105 seems to inhibit growth of #102, an example of an antagonistic interaction. Note that there are sometimes "edge" effects, differences in the appearance of the colony growth in the cultures along the perimeter of the plate as opposed to those growing in a more protected locations (such as the diagonal control colonies). Spend some time with these plates, carefully and fully evaluating all possible combinations of your soil community isolates for as many as possible examples of mutualism or antagonism. Record your results in your lab notebook with copies of the photos of your plates.
If you have positive antibiotic producers or evidence for interesting interactions, please show them to your lab instructor so they can be shared with the class.
Take photos of any plates that show evidence of the presence of antibiotic producers in your soil community. If you found that your isolates did not appear to cause measurable inhibition of growth, does that mean that your isolates do not secrete any antimicrobial compounds? Explain?
Structural Diversity in Cultured Isolates Con't
Complete the Motility Assessment & Analyze the Results
Soft Nutrient Agar Deep Medium
Use Nutrient Agar recipe but reduce agar content to 0.35% Bacteriological Agar. pH 7.6 at 25°C and pour into sterile glass tubes
MOTILITY ASSESSMENT
Look for radiating growth around the stab line of inoculation of each isolate in each of your soft agar deeps. Motility detection is possible due to the semisolid nature (low concentration of agar) of these soft agar deeps. Growth radiating out from the central stab inoculation line indicates that the test organism is motile. First hold an E. coli positive control tube up to the light to see an example of radiating growth. Growth appears cloudier than the medium. Compare your positive control to an uninoculated tube and to a negative control culture of a non-motile organism. Non-motile bacteria exhibit growth in a tighter, defined line limited to where the organism was inoculated. In contrast, motile organisms exhibit detectable growth radiating away from the stab inoculation line towards the periphery. Strictly aerobic organisms may show more growth radiating down from the surface of the medium compared to the growth deep in the tube. Consult with your instructor if you are having a hard time deciding whether or not your isolates are motile. Why might it be useful for some soil community members to be motile?
If you have time, you can try to confirm a positive preliminary motility test by doing a hanging drop motility wet mount or a flagella stain. See the Protocols section in the wiki on Motility Tests for directions on performing confirmation tests.
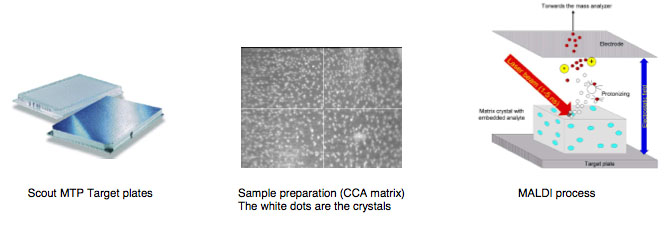
Identification of bacteria using MALDI-TOF Mass Spectronomy
Using Matrix Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry (MALDI-TOF MS) to Identify bacteria
Using log phase (24 hour) cultures of your isolates you will learn how the MALDI-TOF Biotyper works to identify your microbes using a different reference standard than our other ID method (comparison of 16s RNA gene sequences). This method compares particular proteins by mass spectrometry to reference standards. You need freshly grown cultures of your isolates today made 24 hours prior to this lab. If you have one or two very slow growing cultures, you need to allow enough time to have a small but visible colony on your plate.
BACKGROUND:
The following MALDI background information is provided courtesy of Bruker Daltronics:
Glossary
MALDI is the abbreviation for Matrix Assisted Laser Desorption/Ionization
TOF for Time of Flight.
Matrix is a chemical substance which will assist the sample ionization process and must be crystallized in a sample compound on the target surface. Common matrix substances are:
- CCA or HCCA (Alpha-Cyano-4-Hydroxycinnamic Acid)
- SA (Sinapinic Acid)
- DHB (Dihydroxybenzoic Acid)
MALDI-TOF The samples which are measured for MALDI Biotyper application are typically the Proteins of Bacteria, Fungi and Yeast. The mass and distribution information will be used for identification.
By firing with a laser (on microflex a nitrogen uv laser with a wavelength of 337 nm) on the prepared matrix-sample compound the matrix will be vaporized. The sample will be entrained and ionized. The matrix and sample ions will be accelerated in an electrical field as the target plate is under high voltage of up to 20 kV against to an arranged ground potential.
Via the flight time between acceleration and hitting the detector the mass of an ion can be determined (TOF = Time of Flight technique). The complete process needs to take place under vacuum of better than 3*10-6 mbar. As better the vacuum as less collisions
To avoid influences of temperature, mechanical and electrical deviations and special techniques for acceleration the instrument needs to be calibrated before measuring unknown samples. To do this a mixture of known samples will be measured and an assignment between flight time and theoretical mass will be done.
The achievable resolution depends on the speed of the detector and the speed of converting the analog signals to digital signals. On microflex a 500 MHz digitizer which achieves a distance from 2 ns between data points (in 2 ns the light covers a distance of just 60 cm) is used. With a special digitizer function provides actually a calculated ‘pseudo’ 2 GHz sampling rate. Mainly influencing variables are also the applied laser energy, the drift distance and the electrical field distribution.
Definition and calculation of mass resolution
The mass resolution for a single peak is calculated by dividing the peak mass by the Full Width at Half Maximum (FWHM). The resolution value can be checked in flexControl and flexAnalysis directly.
Linear mode
The ionized sample will be accelerated in an electrical field between the target plate and the acceleration electrodes. Followed by ‘drifting’ to a field free region and finished by hitting the detector.
Ion acceleration
Continuous extraction:
The first MALDI-TOFs have been used continuous extraction to accelerate ions. Between target plate and ground plate is a static electrical field which accelerates the ions immediately after generating.
Delayed extraction:
Next step was to use a delayed extraction to accelerate ions (pulsed ion extraction - PIE). Between target plate and ground plate a second potential plate (P2) is placed to modify the electrical field. The P2 plate has the same potential as the target plate. So no acceleration will happen after generating the ions. After a delay time, the P2 potential will be decreased and the acceleration starts. The fortune is that the effect of loosing resolving power because of different kinetic energy of equal ions will be reduced. A higher resolution for a small mass range can be observed.
A further advancement could be reached by optimizing the pulse shape applied to the P2 plate. With the now called PAN (panoramic) mode a high resolution over a brought mass range will be reached.
Electrical Lens:
The ion lens is located on ion optic behind the acceleration electrodes and applies a small vectored electrical field to focus the ion cloud for better resolution.
Connections:
The P1 (IS-1) potential (Target plate) is connected to the PIE electronic J497.
The P2 (IS-2) potential is connected to the PIE electronic J495.
The Lens potential is connected to the Lens power supply J480.
Procedure
The Bruker MALDI Biotyper requires organic acid solutions to: 1) prepare the Bacterial Standard (BTS) and HCCA (matrix): 2) complete the Formic Acid Extraction process; and 3) to clean the Polished Steel Target.
Preparing your isolates for MALDI-TOF MS
Formic Acid Isolate Extraction
Principle: In some microorganisms it may be necessary to break down the cell wall and separate the ribosomal proteins prior to spectra analysis.
Formic Acid Extraction Procedure: Tube Extraction (TE)
Materials Needed :
- Eppendorf 1.5 μL microfuge tubes
- Eppendorf pipettes/tips
- Microfuge
- Toothpicks (or applicable transfer device)
- Vortex
- Microfuge tube rack
- Bacterial Test Standard (BTS)
- Target
- Ultra-Pure Water, HPLC/MS Grade
- Ethanol, 100% HPLC/MS Grade
- Formic Acid, 70% HPLC/MS Grade
- Acetonitrile, 100 % HPLC/MS Grade
- Fresh Microorganism: overnight growth should be used for routine microorganism identification; slow-growing bacteria may need to cultivate for several days before testing; do not use organisms that have been stored at 4°C or lower as this has a negative impact on quality of spectra and reproducibility; storing plates at room temperature for several days is acceptable; different media types and growth temperatures have little effect on results.
- Label a microfuge tube for each of your isolates on the side of the tube using a graphite pencil. Put the code on the top of the tube also using a Sharpie. Use a unique isolate code, for example A2-1 for soil sample and isolate number.
- Add 300 μL of HPLC water to each microfuge tube
- Using a sterile toothpick, or your flame sterilized loop, transfer a visible amount of bacteria from your freshest streak plate to a tube. Twirl the toothpick around in the water to make sure the bacteria are transferred to the water. Discard the tootpick in your autoclave bag.
- Vortex at highest speed for a full minute.
- Check the turbidity of the water in your tube. It should be very cloudy. (You should not be able to see print when you hold up a page of the lab wiki behind it.) If not sufficiently turbid, add more bacteria with another sterile toothpick and vortex again at highest speed for a full minute.
- Add 900 μL of Ethanol
Vortex at highest speed (1) minute.
- Put your tubes in the microcentrifuge and spin them for at maximum speed for (2) minutes
- Ask your instructor to demonstrate the proper way to decant your tubes: take the top off one tube, invert it into the sink and flick a few times to remove as much of the ethanol as possible. BEFORE you revert the tube, touch the lip of the tube to a paper towel to remove the remainder of the ethanol.
- Repeat for each tube.
- Check to make sure you still have a bacterial pellet at the bottom of each tube.
- Use the microcentrifuge to spin all the isolates at highest speed for (2) minutes
- Remove ALL Ethanol in the Speed Vac by spinning for 2 min at high speed with low heat (caps OFF). Repeat if any ethanol or its smell remains in your tubes
- When all traces of ethanol are gone, add 10 μL of 70% Formic Acid.
- Vortex at highest speed for 1 min.
- Let tubes stand for 5 minutes on your bench.
- In the chemical hood and WEARING GLOVES, add 10 μL of 100% Acetonitrile
- Vortex at highest speed for (1) min
- Centrifuge at maximum speed (13,000 to 15,000 rpm) in the microcentrifuge for (2) minute –(continue with: MALDI Target Preparation).
Note: A Bacterial Test Standard (BTS) 10 mg/ml (w/v) is used to calibrate and test software, use fresh on every plate.
The BTS will be provided in lab ready to use.
alpha-Cyano-4-hydroxycinnamic acid (HCCA (MATRIX)) (from Bruker, Fluka, Sigma)
Bruker Product number 255344:.
The reconstituted matrix will be provided in lab ready to use.
MALDI Target Preparation:
Materials Needed:
-
MALDI Biotyper Target
- Bacterial Test Standard (BTS)
- α-Cyano-4-hydroxycinnamic acid HCCA (matrix)
- 2 μL Pipets
- Eppendorf Brand Pipette/Tips
- MALDI Biotyper Target Worksheet
- Fresh Microorganism: overnight growth should be used for routine microorganism identification; slow-growing bacteria may need to cultivate for several days before testing. Do not use organisms that have been stored at 40C or lower, as this has a negative impact on quality of spectra and reproducibility; storing plates at room temperature for several days is acceptable. Different media types and growth temperatures have little effect on results. PREPARE CULTURES FOR MALDI USING FORMIC ACID EXTRACTION IN LAB.
- Bacterial Test Standard (BTS)
TARGET LOADING:
1. Prepare a MALDI Target Worksheet with appropriate sample identification, assigning the first two target spots for BTS (QC) and the rest for each isolate to be analyzed.
2. Inoculate 1 µL of each Isolate Formic Acid extract supernatant (avoid touching the pellet) onto the target spots indicated on the Target Worksheet.
3. Once all the isolate extracts are loaded.
4. One student will place 1μL of BTS on each of the 2 BTS spots.
5. Air Dry all spots at Room temperature.
6. Immediately after BTS spots (and all other spots) are dry, overlay all spots with 1 µL of HCCA (Matrix) (to avoid oxidation of the standard). Once the HCCA is uncapped work quickly to avoid evaporation!!!
7. Be sure the target plate is completely dry before placing the Target plate in the MALDI-TOF.
8. You are ready to analyze your isolates using the MALDI-TOF MS.
Assignment
Graded Assignment:
There is no assignment due in Lab 9:) We will have a workshop on processing and analyzing your culture independent DNA sequencing data. Please bring your laptop computer to lab with you.
There will be a Lab Practical in Lab 10. If you would like to get a head start in studying for it, information can be found at Lab Practical.