BIOL368/F14:Nicole Anguiano Week 8: Difference between revisions
(→Working with Protein Sequences In-class Activity: Added PsiPred and PredictProtein) |
(→Subject Data: changed last no trend sequences) |
||
| (9 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
==Defining Your HIV Structure Research Project== | ==Defining Your HIV Structure Research Project== | ||
Project going to be worked on in conjugation with [[User:Isabel_Gonzaga | Isabel Gonzaga]] and [[User:Chloe_Jones | Chloe Jones]]. The text below is taken from [[BIOL368/F14:Isabel Gonzaga Week 8|Isabel Gonzaga Week 8]], but the project we are working on uses the same question, hypothesis, and subject data. | |||
===Question=== | |||
How does HIV status (diagnosed, progressing or non-trending) affect the structure of the V3 protein region? | |||
===Hypothesis=== | |||
We hypothesize that diagnosed groups will express greater variability in the V3 region in their protein structure, in comparison to the non-trending groups. Initial comparisons show that diagnosed groups and progressing groups expressed greater genetic variability than non-trending groups. These changes may affect the third variable region, affecting the host's ability to adapt to the changes and generate sufficient immune response. | |||
===Subject Data=== | |||
< | According to the [http://bioquest.org/bedrock/problem_spaces/hiv/HIV_data_table_README.pdf BEDROCK HIV Sequence Data Table], I was able to determine which of the subjects used within my study actually developed aids. All 3 AIDS diagnosed were confirmed with the disease by their final visit. In the AIDS progressing groups, subjects developed AIDS within 1 year after their final visit. The Non-Trending groups all maintained high CD4 T Cell Counts above the threshold, even after the study was conducted. Sequences were for each visit and subject were chosen using a [http://www.random.org/integers/ Random Integer Generator], to eliminate selection bias. | ||
<br>The following sequences was taken from the [http://bioquest.org/bedrock/problem_spaces/hiv/nucleotide_sequences.php BEDROCK HIV Problem Space Database], from the Markham et al. (1998) study. <br> | |||
<b>Table 1: Sequences analyzed</b><Br> | |||
{|border="1" | |||
|- | |- | ||
! Group !! Subject !! Visit !! Sequences | ! Group !! Subject !! Visit !! Sequences | ||
|- | |- | ||
| AIDS Diagnosed || 3 <br><br><br>10<br><br><br>15|| 1<br>6 <br><Br>1<br>6<br><br>1<br>4|| 1, 2, | | AIDS Diagnosed || 3 <br><br><br>10<br><br><br>15|| 1<br>6 <br><Br>1<br>6<br><br>1<br>4|| 1, 2, 4<br>3, 4, 5<br><br>3, 6, 7<br>2, 4, 8<br><br>2, 3, 4<br> 5, 8, 10 | ||
|- | |- | ||
| AIDS Progressing || | | AIDS Progressing || 7 <br><br><br>8<br><br><br>14|| 1<br>5<br><Br>1<br>7<br><br>1<br>9|| 2, 3, 9<br>2, 8, 9<br><br>1, 4, 5<br>1, 6, 7<br><br>2, 3, 4<br>9, 10, 11 | ||
|- | |- | ||
| No Trend || 5 <br><br><br>6<br><br><br>13|| 1<br>5 <br><Br>1<br> | | No Trend || 5 <br><br><br>6<br><br><br>13|| 1<br>5 <br><Br>1<br>9<br><br>1<br>5|| 1, 3, 8<br>4, 5, 2<br><br>1, 2, 3<br>6, 7, 9<br><br>1, 3, 4<br> 3, 5, 4 | ||
|} | |} | ||
==Working with Protein Sequences In-class Activity== | ==Working with Protein Sequences In-class Activity== | ||
| Line 88: | Line 89: | ||
===Working with a single protein sequence=== | ===Working with a single protein sequence=== | ||
====ProtParam==== | ====ProtParam==== | ||
*I navigated to [http://web.expasy.org/protparam/ ProtParam], and inputted the sequence from the | *I navigated to [http://web.expasy.org/protparam/ ProtParam], and inputted the sequence from the clone above, then selected "Compute Parameters". The result was as follows: | ||
<b>Number of amino acids</b>: | <b>Number of amino acids</b>: 94 <br /> | ||
<b>Molecular weight</b>: | <b>Molecular weight</b>: 10625.1 <br /> | ||
<b>Theoretical pI</b>: | <b>Theoretical pI</b>: 10.14 <br /><br /> | ||
<b>Amino acid composition</b>: | <b>Amino acid composition</b>: | ||
{|border="1" | {|border="1" | ||
| Ala (A) || | | Ala (A) || 2 || 2.1% | ||
|- | |- | ||
| | | Arg (R) || 6 || 6.4% | ||
|- | |- | ||
| | | Asn (N) || 14 || 14.9% | ||
|- | |- | ||
| | | Asp (D) || 1 || 1.1% | ||
|- | |- | ||
| | | Cys (C) || 2 || 2.1% | ||
|- | |- | ||
| | | Gln (Q) || 4 || 4.3% | ||
|- | |- | ||
| | | Glu (E) || 4 || 4.3% | ||
|- | |- | ||
| | | Gly (G) || 5 || 5.3% | ||
|- | |- | ||
| | | His (H) || 2 || 2.1% | ||
|- | |- | ||
| | | Ile (I) || 14 || 14.9% | ||
|- | |- | ||
| | | Leu (L) || 3 || 3.2% | ||
|- | |- | ||
| Met (M) || | | Lys (K) || 6 || 6.4% | ||
|- | |||
| Met (M) || 0 || 0.0% | |||
|- | |||
| Phe (F) || 4 || 4.3% | |||
|- | |||
| Pro (P) || 3 || 3.2% | |||
|- | |||
| Ser (S) || 8 || 8.5% | |||
|- | |- | ||
| | | Thr (T) || 7 || 7.4% | ||
|- | |||
| Trp (W) || 1 || 1.1% | |||
|- | |||
| Tyr (Y) || 1 || 1.1% | |||
|- | |||
| Val (V) || 7 || 7.4% | |||
|- | |- | ||
| | | Pyl (O) || 0 || 0.0% | ||
|- | |||
| Sec (U) || 0 || 0.0% | |||
|- | |||
| | |||
|- | |- | ||
| | | (B) || 0 || 0.0% | ||
|- | |- | ||
| ( | | (Z) || 0 || 0.0% | ||
|- | |- | ||
| ( | | (X) || 0 || 0.0% | ||
|}<br /> | |||
|} <br /> | |||
<b>Total number of negatively charged residues (Asp + Glu)</b>: | <b>Total number of negatively charged residues (Asp + Glu)</b>: 5 <br /> | ||
<b>Total number of positively charged residues (Arg + Lys)</b>: | <b>Total number of positively charged residues (Arg + Lys)</b>: 12 <br /><br /> | ||
<b>Atomic composition</b>: | <b>Atomic composition</b>: | ||
{|border="1" | {|border="1" | ||
|Carbon || C || | |Carbon || C || 466 | ||
|- | |- | ||
| Hydrogen || H || | | Hydrogen || H || 759 | ||
|- | |- | ||
| Nitrogen || N || | | Nitrogen || N || 141 | ||
|- | |- | ||
| Oxygen || O || | | Oxygen || O || 139 | ||
|- | |- | ||
| Sulfur || S || | | Sulfur || S || 2 | ||
|} <br /> | |} <br /> | ||
<b>Formula</b>: C<sub> | <b>Formula</b>: C<sub>466</sub>H<sub>759</sub>N<sub>141</sub>O<sub>139</sub>S<sub>2</sub> <br /> | ||
<b>Total number of atoms</b>: 13540 <br /> <br /> | <b>Total number of atoms</b>: 13540 <br /> <br /> | ||
<b>Extinction coefficients</b>:<br /> | <b>Extinction coefficients</b>:<br /> | ||
Extinction coefficients are in units of M-1 cm-1, at 280 nm measured in water. <br /> | Extinction coefficients are in units of M-1 cm-1, at 280 nm measured in water. <br /> | ||
Ext. coefficient | Ext. coefficient 7115 <br /> | ||
Abs 0.1% (=1 g/l) | Abs 0.1% (=1 g/l) 0.670, assuming all pairs of Cys residues form cystines<br /> | ||
Ext. coefficient | Ext. coefficient 6990<br /> | ||
Abs 0.1% (=1 g/l) | Abs 0.1% (=1 g/l) 0.658, assuming all Cys residues are reduced<br /><br /> | ||
<b>Estimated half-life</b>:<br /> | <b>Estimated half-life</b>:<br /> | ||
The N-terminal of the sequence considered is M (Met).<br /> | The N-terminal of the sequence considered is M (Met).<br /> | ||
The estimated half-life is: | The estimated half-life is: 1 hours (mammalian reticulocytes, in vitro), 30 min (yeast, in vivo), >10 hours (Escherichia coli, in vivo).<br /><br /> | ||
<b>Instability index</b>:<br /> | <b>Instability index</b>:<br /> | ||
The instability index (II) is computed to be | The instability index (II) is computed to be 45.96<br /> | ||
This classifies the protein as | This classifies the protein as unstable.<br /> | ||
<b>Aliphatic index</b>: | <b>Aliphatic index</b>: 94.26<br /> | ||
<b>Grand average of hydropathicity (GRAVY)</b>: -0. | <b>Grand average of hydropathicity (GRAVY)</b>: -0.362 | ||
====ProtScale==== | ====ProtScale==== | ||
*I navigated to [http://web.expasy.org/protscale/ ProtScale] and entered the amino acid sequence. I changed the "Window Size" dropdown to 19, then hit Submit. I saved the image as a .gif (Fig. 2). | *I navigated to [http://web.expasy.org/protscale/ ProtScale] and entered the amino acid sequence. I changed the "Window Size" dropdown to 19, then hit Submit. I saved the image as a .gif (Fig. 2). | ||
[[Image: | [[Image:S15V4C3ProtScale.gif|thumb|none|upright=2|alt=Protscale result for subject 15, visit 4, clone 3's amino acid sequence.|<b>Figure 2</b>: The ProtScale result for subject 15, visit 4, clone 3's amino acid sequence..]] | ||
====TMHMM==== | ====TMHMM==== | ||
*Next, I navigated to [http://www.cbs.dtu.dk/services/TMHMM/ TMHMM], pasted in the sequence, then hit submit, then saved the image (Fig. 3). | *Next, I navigated to [http://www.cbs.dtu.dk/services/TMHMM/ TMHMM], pasted in the sequence, then hit submit, then saved the image (Fig. 3). | ||
[[Image: | [[Image:S14V4C3TMHMM.png|thumb|none|upright=2|alt=TMHMM result for subject 15, visit 4, clone 3's amino acid sequence.|<b>Figure 3</b>: The TMHMM result for subject 15, visit 4, clone 3's amino acid sequence. Note the lack of any visible lines.]] | ||
====ScanProsite==== | ====ScanProsite==== | ||
| Line 202: | Line 203: | ||
*I navigated to [http://www.ebi.ac.uk/Tools/pfa/iprscan5/ InterProScan] and inputted the amino acid sequence and hit submit. | *I navigated to [http://www.ebi.ac.uk/Tools/pfa/iprscan5/ InterProScan] and inputted the amino acid sequence and hit submit. | ||
[[Image:InterProScanS15V4C3.png|thumb|none|upright=3|alt=InterProScan5 Results|<b>Figure 6</b>: The InterProScan results showing the predicted domains of the protein.]] | [[Image:InterProScanS15V4C3.png|thumb|none|upright=3|alt=InterProScan5 Results|<b>Figure 6</b>: The InterProScan results showing the predicted domains of the protein. The results show the protein to be a member of gp160.]] | ||
====CD Server==== | ====CD Server==== | ||
| Line 216: | Line 217: | ||
*I navigated to [http://bioinf.cs.ucl.ac.uk/psipred/ PsiPred]. I inputted the amino acid sequence and gave it the identifier "S15V4C3", then hit Predict. I waited about 15 minutes until it finished the prediction. | *I navigated to [http://bioinf.cs.ucl.ac.uk/psipred/ PsiPred]. I inputted the amino acid sequence and gave it the identifier "S15V4C3", then hit Predict. I waited about 15 minutes until it finished the prediction. | ||
[[Image:PsiPredS15V4C3.png|thumb|none|upright=2|alt= | [[Image:PsiPredS15V4C3.png|thumb|none|upright=2|alt=PsiPred result|<b>Figure 8</b>: The results from PsiPred using the amino acid from Subject 15, visit 4, clone 3. Note the two alpha helices and presence of many beta sheets.]] | ||
====PredictProtein==== | ====PredictProtein==== | ||
*I navigated to [https://www.predictprotein.org/ Predict Protein]. I created an account so I could utilize the service. Then I validated my account and returned to the site. I logged in and inputted the amino acid sequence. I then resubmitted the job to get current results. | *I navigated to [https://www.predictprotein.org/ Predict Protein]. I created an account so I could utilize the service. Then I validated my account and returned to the site. I logged in and inputted the amino acid sequence. I then resubmitted the job to get current results. The detailed results are visible [https://www.predictprotein.org/get_results?req_id=487817 here]. | ||
[[Image:PredictProteinS15V4C3Old.png|thumb|none|upright=3|alt=PredictProtein Old results|<b>Figure 9</b>: The results using the autogenerated PredictProtein results. The red bars are alpha helices.]] | |||
[[Image:PredictProteinS15V4C3New.png|thumb|none|upright=3|alt=PredictProtein New results|<b>Figure 10</b>:The results using the newly generated PredictProtein results. The red bars are alpha helices. Note the one large alpha helix and the one smaller one.]] | |||
===Crystal Structure Comparison=== | |||
*I navigated to [http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?uid=2B4C NCBI] and downloaded the structure as a CN3D file. I opened the file in CN3D (Fig. 11), and selected the amino acid sequence that corresponded to the similar sequence to what Translate returned on the given amino acid sequence (Fig. 1). I selected "Show Selected Residues" to display only what was selected (Fig. 12). The presence of a smaller alpha helix in both PsiPred (Fig. 8) and PredictProtein (Fig. 10) indicates that mutations in the protein may have caused an alpha helix to form. However, the one large alpha helix is likely the alpha helix present in the original crystal structure. The presence of many beta sheets goes alongside the presence of beta sheets as seen from PsiPref (Fig. 8). | |||
[[Image:Gp120crystalstructureCN3D.png|thumb|none|upright=4|alt=gp120 crystal structure|<b>Figure 11</b>: The crystal structure of gp120.]] | |||
[[Image:CN3DSelectedRegionS15.png|thumb|none|upright=2|alt=selected amino acid sequence in gp120|<b>Figure 12</b>: The protein that is coded for by the amino acid sequence in gp120 that is closest to the amino acid sequence returned by subject 15, visit 4, clone 3.]] | |||
==Links== | ==Links== | ||
{{Template:Nicole Anguiano}} | {{Template:Nicole Anguiano}} | ||
Latest revision as of 14:45, 22 October 2014
Defining Your HIV Structure Research Project
Project going to be worked on in conjugation with Isabel Gonzaga and Chloe Jones. The text below is taken from Isabel Gonzaga Week 8, but the project we are working on uses the same question, hypothesis, and subject data.
Question
How does HIV status (diagnosed, progressing or non-trending) affect the structure of the V3 protein region?
Hypothesis
We hypothesize that diagnosed groups will express greater variability in the V3 region in their protein structure, in comparison to the non-trending groups. Initial comparisons show that diagnosed groups and progressing groups expressed greater genetic variability than non-trending groups. These changes may affect the third variable region, affecting the host's ability to adapt to the changes and generate sufficient immune response.
Subject Data
According to the BEDROCK HIV Sequence Data Table, I was able to determine which of the subjects used within my study actually developed aids. All 3 AIDS diagnosed were confirmed with the disease by their final visit. In the AIDS progressing groups, subjects developed AIDS within 1 year after their final visit. The Non-Trending groups all maintained high CD4 T Cell Counts above the threshold, even after the study was conducted. Sequences were for each visit and subject were chosen using a Random Integer Generator, to eliminate selection bias.
The following sequences was taken from the BEDROCK HIV Problem Space Database, from the Markham et al. (1998) study.
Table 1: Sequences analyzed
| Group | Subject | Visit | Sequences |
|---|---|---|---|
| AIDS Diagnosed | 3 10 15 |
1 6 1 6 1 4 |
1, 2, 4 3, 4, 5 3, 6, 7 2, 4, 8 2, 3, 4 5, 8, 10 |
| AIDS Progressing | 7 8 14 |
1 5 1 7 1 9 |
2, 3, 9 2, 8, 9 1, 4, 5 1, 6, 7 2, 3, 4 9, 10, 11 |
| No Trend | 5 6 13 |
1 5 1 9 1 5 |
1, 3, 8 4, 5, 2 1, 2, 3 6, 7, 9 1, 3, 4 3, 5, 4 |
Working with Protein Sequences In-class Activity
Reading a SWISS-PROT Entry
- I navigated to UniProt. I searched for "Q75760". Here is a portion of the results from the protein that came up from the search.
Entry Information
Entry Name: Q75760_9HIV1
Primary (citable) accession number: Q75760
Integrated into UniProtKB/TrEMBL: November 1, 1996
Last sequence update: November 1, 1996
Last modified: October 1, 2014
Names & Taxonomy
Protein names: Envelope glycoprotein gp160
Gene names: env
Organism: Human immunodeficiency virus 1
Taxonomic identifier: 11676
Taxonomic lineage: Viruses › Retro-transcribing viruses › Retroviridae › Orthoretrovirinae › Lentivirus › Primate lentivirus group
Function
The envelope glyprotein gp160 precursor down-modulates cell surface CD4 antigen by interacting with it in the endoplasmic reticulum and blocking its transport to the cell surface.
The gp120-gp41 heterodimer allows rapid transcytosis of the virus through CD4 negative cells such as simple epithelial monolayers of the intestinal, rectal and endocervical epithelial barriers. Both gp120 and gp41 specifically recognize glycosphingolipids galactosyl-ceramide (GalCer) or 3' sulfo-galactosyl-ceramide (GalS) present in the lipid rafts structures of epithelial cells. Binding to these alternative receptors allows the rapid transcytosis of the virus through the epithelial cells. This transcytotic vesicle-mediated transport of virions from the apical side to the basolateral side of the epithelial cells does not involve infection of the cells themselves.
Interaction
Binary Interactions
| With | Entry | #Exp | IntAct | Notes |
|---|---|---|---|---|
| P84801 | 2 | EBI-8453491,EBI-8453570 | From a different organism. | |
| ath | Q9KWN0 | 2 | EBI-8453491,EBI-8453511 | From a different organism. |
| UDA1 | P11218 | 2 | EBI-8453491,EBI-8453649 | From a different organism. |
Protein-protein interaction databases
| Dip | DIP-59960N. |
| IntAct | Q75760. 3 interactions. |
| MINT | MINT-8414778. |
Subcellular Location
Virion membrane; Single-pass type I membrane protein. Host cell membrane; Single-pass type I membrane protein. Host endosome membrane; Single-pass type I membrane protein
PTM / Processing
Amino Acid modifications
| Feature Key | Position(s) | Length | Description |
|---|---|---|---|
| Glycosylation | 298-298 | 1 | N-linked (GlcNAc...) |
Miscellaneous
Keywords - Technical term
3D-structure
Cross-References
- This section contained a variety of references. There were sequence databases (EMBL, GenBank, DDBJ, PIR), 3D structure databases (PDBe, RCSB PDB, PDBj, ProteinModelPortal, SMR, ModBase, ModiDB), protein-protein interaction databases (DIP, IntAct, MINT), protocols and materials databases (Structural Biology Knowledgebase), miscellaneous databases (EvolutionaryTrace), and family and domain databases (Gene3D, InterPro, Pfam, SUBFAM, Protonet).
Features
See table under PTM / Processing.
Question
- If you search on the keywords "HIV" and "gp120", how many results do you get?
- Searching "hiv" returns 600,415 results. Searching "gp120" returns 182,286 results. Searching "hiv AND gp120" returned 180,227 results.
ORFing your DNA sequence
- I chose to ORF the DNA sequence from subject 15, visit 4, clone 3. Using Translate, I inputted the DNA sequence and obtained the 6 possible open reading frames.

- Comparing to the fasta sequence of the Uniprot protein above, I can see that the first open reading frame is most likely the first. The amino acid sequence, "EVVIRSENFTNNAKIIIVHLNESVVINCTRPNNNTRRKIPIGPGSSFYTTGIIGDIRQAHCNISGSKWNNTLKQIVNKLREQFVNKTIIFNQSS", is extremely similar to the sequence contained in the Uniprot protein, "EVVIRSDNFTNNAKTIIVQLKESVEINCTRPNNNTRKSIHIGPGRAFYTTGEIIGDIRQAHCNISRAKWNDTLKQIVIKLREQFENKTIVFNHSS". There are very few differences between them, indicating that likely the env gene is located in that location in the overall protein.
Working with a single protein sequence
ProtParam
- I navigated to ProtParam, and inputted the sequence from the clone above, then selected "Compute Parameters". The result was as follows:
Number of amino acids: 94
Molecular weight: 10625.1
Theoretical pI: 10.14
Amino acid composition:
| Ala (A) | 2 | 2.1% |
| Arg (R) | 6 | 6.4% |
| Asn (N) | 14 | 14.9% |
| Asp (D) | 1 | 1.1% |
| Cys (C) | 2 | 2.1% |
| Gln (Q) | 4 | 4.3% |
| Glu (E) | 4 | 4.3% |
| Gly (G) | 5 | 5.3% |
| His (H) | 2 | 2.1% |
| Ile (I) | 14 | 14.9% |
| Leu (L) | 3 | 3.2% |
| Lys (K) | 6 | 6.4% |
| Met (M) | 0 | 0.0% |
| Phe (F) | 4 | 4.3% |
| Pro (P) | 3 | 3.2% |
| Ser (S) | 8 | 8.5% |
| Thr (T) | 7 | 7.4% |
| Trp (W) | 1 | 1.1% |
| Tyr (Y) | 1 | 1.1% |
| Val (V) | 7 | 7.4% |
| Pyl (O) | 0 | 0.0% |
| Sec (U) | 0 | 0.0% |
| (B) | 0 | 0.0% |
| (Z) | 0 | 0.0% |
| (X) | 0 | 0.0% |
Total number of negatively charged residues (Asp + Glu): 5
Total number of positively charged residues (Arg + Lys): 12
Atomic composition:
| Carbon | C | 466 |
| Hydrogen | H | 759 |
| Nitrogen | N | 141 |
| Oxygen | O | 139 |
| Sulfur | S | 2 |
Formula: C466H759N141O139S2
Total number of atoms: 13540
Extinction coefficients:
Extinction coefficients are in units of M-1 cm-1, at 280 nm measured in water.
Ext. coefficient 7115
Abs 0.1% (=1 g/l) 0.670, assuming all pairs of Cys residues form cystines
Ext. coefficient 6990
Abs 0.1% (=1 g/l) 0.658, assuming all Cys residues are reduced
Estimated half-life:
The N-terminal of the sequence considered is M (Met).
The estimated half-life is: 1 hours (mammalian reticulocytes, in vitro), 30 min (yeast, in vivo), >10 hours (Escherichia coli, in vivo).
Instability index:
The instability index (II) is computed to be 45.96
This classifies the protein as unstable.
Aliphatic index: 94.26
Grand average of hydropathicity (GRAVY): -0.362
ProtScale
- I navigated to ProtScale and entered the amino acid sequence. I changed the "Window Size" dropdown to 19, then hit Submit. I saved the image as a .gif (Fig. 2).

TMHMM
- Next, I navigated to TMHMM, pasted in the sequence, then hit submit, then saved the image (Fig. 3).

ScanProsite
- I navigated to ScanProsite and inputted the amino acid sequence. I deselected "Exclude motifs with a high probability of occurrence from the scan", and then hit "START THE SCAN".

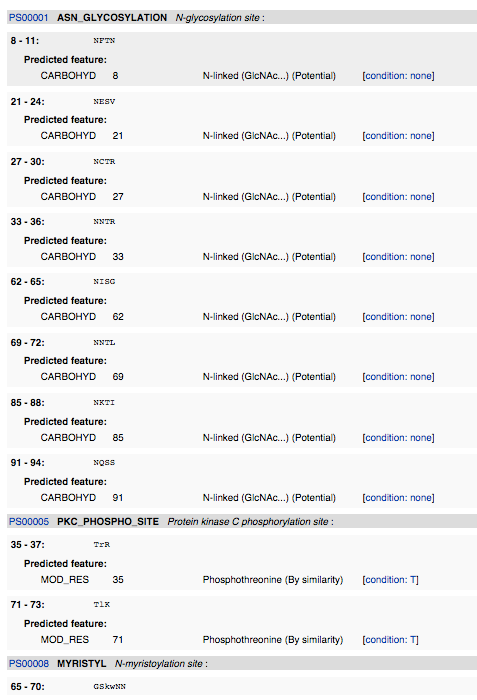
InterProScan
- I navigated to InterProScan and inputted the amino acid sequence and hit submit.

CD Server
- I navigated to CD Server and inputted the amino acid sequence. Then I changed the Expect Value Threshlod to 1, and hit submit.

Predicting the Secondary Structure of a Protein
PsiPred
- I navigated to PsiPred. I inputted the amino acid sequence and gave it the identifier "S15V4C3", then hit Predict. I waited about 15 minutes until it finished the prediction.

PredictProtein
- I navigated to Predict Protein. I created an account so I could utilize the service. Then I validated my account and returned to the site. I logged in and inputted the amino acid sequence. I then resubmitted the job to get current results. The detailed results are visible here.


Crystal Structure Comparison
- I navigated to NCBI and downloaded the structure as a CN3D file. I opened the file in CN3D (Fig. 11), and selected the amino acid sequence that corresponded to the similar sequence to what Translate returned on the given amino acid sequence (Fig. 1). I selected "Show Selected Residues" to display only what was selected (Fig. 12). The presence of a smaller alpha helix in both PsiPred (Fig. 8) and PredictProtein (Fig. 10) indicates that mutations in the protein may have caused an alpha helix to form. However, the one large alpha helix is likely the alpha helix present in the original crystal structure. The presence of many beta sheets goes alongside the presence of beta sheets as seen from PsiPref (Fig. 8).


Links
Nicole Anguiano
BIOL 368, Fall 2014
Assignment Links
- Week 1 Assignment
- Week 2 Assignment
- Week 3 Assignment
- Week 4 Assignment
- Week 5 Assignment
- Week 6 Assignment
- Week 7 Assignment
- Week 8 Assignment
- Week 9 Assignment
- Week 10 Assignment
- Week 11 Assignment
- Week 12 Assignment
- Week 13 Assignment
- Week 15 Assignment
Individual Journals
- Individual Journal Week 2
- Individual Journal Week 3
- Individual Journal Week 4
- Individual Journal Week 5
- Individual Journal Week 6
- Individual Journal Week 7
- Individual Journal Week 8
- Individual Journal Week 9
- Individual Journal Week 10
- Individual Journal Week 11
- Individual Journal Week 12
- Individual Journal Week 13
- Individual Journal Week 15