BIOL368/F14:Nicole Anguiano Week 5: Difference between revisions
(→Conclusions: added in a blurb about how none of my week 3 reviews covered this) |
(→Electronic Lab Notebook: moved link so it's more visible and edited it so it would immediately download instead of making you go to another page) |
||
| (One intermediate revision by the same user not shown) | |||
| Line 1: | Line 1: | ||
==Electronic Lab Notebook== | ==Electronic Lab Notebook== | ||
[[Media:NicoleAnguiano_ResearchPresentation.pptx | Presentation link.]] | |||
Answering the question: Do clones from a particular subject's group (rapidly progressing, moderately progressing, and nonprogressing), share any genetic similarity with one another? Are clones from subjects in the same group the most similar, or are they as dissimilar as clones from other groups? Does the amount of variation and similarity, if any, change from the time of the first visit to a visit after about 2 years of infection with the virus? | Answering the question: Do clones from a particular subject's group (rapidly progressing, moderately progressing, and nonprogressing), share any genetic similarity with one another? Are clones from subjects in the same group the most similar, or are they as dissimilar as clones from other groups? Does the amount of variation and similarity, if any, change from the time of the first visit to a visit after about 2 years of infection with the virus? | ||
Latest revision as of 00:04, 1 October 2014
Electronic Lab Notebook
Answering the question: Do clones from a particular subject's group (rapidly progressing, moderately progressing, and nonprogressing), share any genetic similarity with one another? Are clones from subjects in the same group the most similar, or are they as dissimilar as clones from other groups? Does the amount of variation and similarity, if any, change from the time of the first visit to a visit after about 2 years of infection with the virus?
Below are the clones chosen and the subjects/visits they are from:
| Progressor Group | Subject | Visit # | Clones | Visit # | Clones |
|---|---|---|---|---|---|
| Rapid | 3 | 1 | 1, 2, 3 | 3 | 2, 7, 9 |
| Rapid | 11 | 1 | 3, 5, 7 | 3 | 3, 6, 9 |
| Rapid | 15 | 1 | 3, 6, 12 | 4 | 1, 3, 4 |
| Moderate | 6 | 1 | 1, 2, 3 | 5 | 2, 4, 8 |
| Moderate | 8 | 1 | 1, 3, 5 | 4 | 4, 5, 6 |
| Moderate | 14 | 1 | 2, 3, 6 | 5 | 1, 6, 7 |
| Nonprogressor | 2 | 1 | 3, 4, 5 | 3 | 5, 6, 7 |
| Nonprogressor | 12 | 1 | 1, 2, 3 | 4 | 2, 5, 10 |
| Nonprogressor | 13 | 1 | 1, 3, 4 | 3 | 1, 2, 4 |
Methods and Results
- First, I compiled the sequences of each of the clones that I'd specified last week (also listed above) from the bioquest web site. I went to the Biology workbench and created a new session, called "Research". In this session, I uploaded the file I just created with all of the sequences.
Visit 1
- I began by running a CLUSTALW on each of the first visit clones. I ran one on the rapid progressors, then the moderate progressors, then the nonprogressors, importing the alignments each time so that further analysis could be performed on each alignment through CLUSTALDIST.


- The rapid progressors have a total of 85 differences between their collective sequences (S=85, Table 1). Each progressor is most similar to their own clones, as expected, although clone 6 from subject 15 is quite different from the other clones from subject 15. It is possible that the S value is increased due to the large number of differences between subject 15 clone 6 and the rest of the subject 15 clones. The rapid progressors are notable for being the only group in which more than one subject has additional nucleotides not present in the rest in more than one place. This could also be a cause of the very high S value.

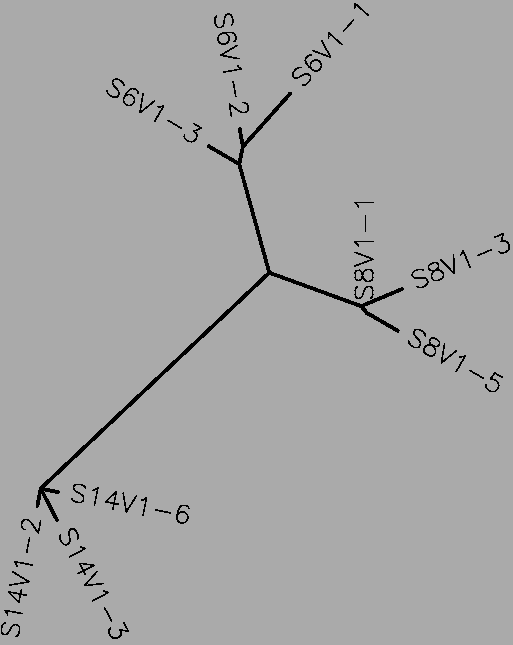
- The moderate progressors have a total of 49 differences between their collective sequences (S=49, Table 1). Each progressor is most similar to its own clones, as expected. No one subject has a particularly different clone, unlike the rapid progressors (Fig. 2). Despite also having one area in which one subject has additional nucleotides, the S value is relatively low.


- The nonprogressors have a total of 55 differences between their collective sequences (S=55, Table 1). Each progressor is most similar to its own clones, as expected. No one subject has a particularly different clone, unlike the rapid progressors. There are no areas in which a clone has extra nucleotides or missing nucleotides, so the differences are only in the base 288 bases.
- After running the CLUSTALW, I ran a CLUSTALDIST on each group to find the clustal distance matrix, and calculate the theta, min difference, and max difference (Table 1).

- The minimum value is 0.135, and the maximum is 0.177. This is used to calculate the minimum and maximum differences in Table 1 using the gene length of 291.

- The minimum value is 0.049, and the maximum is 0.112. This is used to calculate the minimum and maximum differences in Table 1 using the gene length of 288.

- The minimum value is 0.077, and the maximum is 0.154. This is used to calculate the minimum and maximum differences in Table 1 using the gene length of 285.
| Group | S | θ | Min Difference | Max Difference |
|---|---|---|---|---|
| Rapid Progressors | 85 | [math]\displaystyle{ \textstyle\frac{170}{3} }[/math] | 39 | 52 |
| Moderate Progressors | 49 | [math]\displaystyle{ \textstyle\frac{98}{3} }[/math] | 14 | 32 |
| Nonprogressors | 55 | [math]\displaystyle{ \textstyle\frac{110}{3} }[/math] | 22 | 44 |
- Table 1: The S, θ, and minimum/maximum differences among the individual progressor groups. Unusually, the moderate progressors are actually the most similar group, the the rapid progressors predictably having the most differences between them. It may have been predicted that the nonprogressors had the most differences, but the moderate progressors came out the most similar, with an the maximum difference of the nonprogressors being 12 over the maximum difference of the moderate progressors.
- After comparing each group with itself, I then compared across groups. I ran a CLUSTALW and CLUSTALDIST on the first visit clones from the three rapid progressors and the three moderate progressors, then the three rapid progressors and the three nonprogressors, then the three moderate progressors and the three nonprogressors. After the comparison, I ran a CLUSTALW and CLUSTALDIST on all of the visit 1 clones.



- There is actually not as large of a difference between the rapid and moderate progressors as would have been expected. Subjects 14 and 15 seem more similar than either of their rapid or moderate counterparts, respectively. Subject 11 remains vastly different than the other two, while subjects 8 and 6 are relatively similar. The lowest value in the clustal distance matrix was 0.064, and the highest was 0.191, which was used to calculate the min and max differences in table 2. Comparisons across the clustal distance matrix was done between groups and not within them, so only rapid progressors were compared with moderate progressors and moderate progressors were compared with rapid progressors. This comparison scheme will remain for the remainder of the clustal distance matrices.



- As may have been expected, the rapid and nonprogeressors are quite different, much moreso than the moderate and rapid progressors. No close similarities exist like in subjects 14 and 15 (Fig. 11). The nonprogressors appear different from both each other and the rapid progressors, and the rapid progressors appear both different from each other and the nonprogressors. The lowest value in the clustal distance matrix was 0.082, and the highest was 0.186, which was used to calculate the min and max differences in table 2.



- Subject 6 and 8 from the moderate progressors are relatively similar, but there do not exist any close similarities between the moderate and the nonprogressors. Overall, the moderate and nonprogressors are relatively different and don't share a large amount of differences. The lowest value in the clustal distance matrix was 0.074, and the highest was 0.165, which was used to calculate the min and max differences in table 2.


- Unusually, the two most similar subjects were 14 and 15, one of which was a moderate progressor and one of which was a rapid progressor, respectively. The next most similar are 6 and 8, both moderate progressors. Outside of those, the subjects were all relatively different. The lowest value in the clustal distance matrix was 0.071 and the highest was 0.186.
| Groups Being Compared | Min Difference | Max Difference |
|---|---|---|
| Rapid and Moderate | 19 | 56 |
| Rapid and Nonprogressor | 24 | 54 |
| Moderate and Nonprogressor | 21 | 48 |
| All | 21 | 54 |
- Table 2: The minimum and maximum differences between the progressor groups for the first visit. Strangely, the rapid and moderate groups have both the lowest minimum difference and the highest maximum difference, indicating the largest range of difference between the two of them. The rapid progressors are more similar to the moderate progressors than the are to each other, having a lower minimum difference between the two than just within the rapid progressors. However, they also have a higher maximum difference, indicating that there are clones that are more varied. The rapid progressors and the nonprogressors are more different than either of their two respective groups, though again the minimum difference is lower than it is just among the rapid progressors. The moderate progressors and nonprogressors are more different overall, with the minimum difference being only one lower than the minimum difference of the nonprogressors.
Mid-Visit
- I moved from the first visit clones to the mid-visit clones, and ran a CLUSTALW on each of the mid-visit clones. I ran one on the rapid progressors, then the moderate progressors, then the nonprogressors, importing the alignments each time so that further analysis could be performed on each alignment through CLUSTALDIST.


- The rapid progressors have a total of 83 differences between their collective sequences (S=83, Table 3). Each progressor is most similar to their own clones, as expected, with the three clones from subject 11 being extremely similar. Unlike the first visit, none of the subjects have one clone that is very different from the rest. This removes the possibility that the extreme difference of one clone is a significant cause of the high S value, and indicates the the rapid progressors are simply more different than one another than the rest of the groups.


- The moderate progressors have a total of 50 differences between their collective sequences (S=50, Table 3). Each progressor is most similar to their own clones, as expected. Clones 4 and 8 from subject 6 are more different than the rest (as well as more different than what was seen in the visit 1 clones), but not significantly.


- The moderate progressors have a total of 50 differences between their collective sequences (S=50, Table 3). Each progressor is most similar to their own clones, as expected. The subject 13 clones are extremely similar, as are the clones of subject 12, with the clones of subject 2 being more diverse than the rest. However, overall, they are all quite similar.
- After running the CLUSTALW, I ran a CLUSTALDIST on each group to find the clustal distance matrix, and calculate the theta, min difference, and max difference (Table 3).

- The minimum value is 0.142, and the maximum is 0.177. This is used to calculate the minimum and maximum differences in Table 3 using the gene length of 291.

- The minimum value is 0.042, and the maximum is 0.112. This is used to calculate the minimum and maximum differences in Table 3 using the gene length of 288.

- The minimum value is 0.077, and the maximum is 0.158. This is used to calculate the minimum and maximum differences in Table 3 using the gene length of 285.
| Group | S | θ | Min Difference | Max Difference |
|---|---|---|---|---|
| Rapid Progressors | 83 | [math]\displaystyle{ \textstyle\frac{166}{3} }[/math] | 41 | 52 |
| Moderate Progressors | 50 | [math]\displaystyle{ \textstyle\frac{100}{3} }[/math] | 12 | 32 |
| Nonprogressors | 58 | [math]\displaystyle{ \textstyle\frac{116}{3} }[/math] | 22 | 45 |
- Table 3: The S, θ, and minimum/maximum differences among the individual progressor groups. Again, the moderate progressors are the most similar of the group. The rapid progressors are the most diffierent, with the minimum and maximum differences only differing by a value of 9. The nonprogressors, again, are in the middle, but have the largest range, with the minimum and maximum differences differing by 23.
- After comparing each group with itself, I then compared across groups. I ran a CLUSTALW and CLUSTALDIST on the mid-visit clones from the three rapid progressors and the three moderate progressors, then the three rapid progressors and the three nonprogressors, then the three moderate progressors and the three nonprogressors. After the comparison, I ran a CLUSTALW and CLUSTALDIST on all of the mid-visit clones.



- Again, subjects 14 and 15 seem more similar than either of their rapid or moderate counterparts, respectively. Subjects 8 and 6 are relatively similar, again. Subjects 3 and 11 are quite different from all the subjects. The lowest value in the clustal distance matrix was 0.060, and the highest was 0.184, which was used to calculate the min and max differences in table 4.



- It doesn't seem as though there are any significant similarities between and of the rapid progressors or nonprogressors. Subjects 2 and 15 are the most similar among the groups, but the similarity is relatively small. The lowest value in the clustal distance matrix was 0.078, and the highest was 0.179, which was used to calculate the min and max differences in table 4.



- While the moderate and nonprogressors seem more closely related than the rapid and nonprogressors, there still are no significant simiarities. The most closely related are subjects 6 and 8, from the moderate progressors. The lowest value in the clustal distance matrix was 0.067, and the highest was 0.158, which was used to calculate the min and max differences in table 4.


- Subjects 6 and 8, as well as subjects 14 and 15, were again the most similar. The former two were from the moderate progressors, and the latter two were from the moderate and rapid progressor groups respectively. The lowest value in the clustal distance matrix was 0.070 and the highest was 0.179, which was used to calculate the min and max differences in table 4.
| Groups Being Compared | Min Difference | Max Difference |
|---|---|---|
| Rapid and Moderate | 17 | 54 |
| Rapid and Nonprogressor | 23 | 52 |
| Moderate and Nonprogressor | 19 | 46 |
| All | 20 | 52 |
- Table 4: The minimum and maximum differences between the progressor groups for the middle visit. The rapid and moderate groups have both the lowest minimum difference and the highest maximum difference again. The trends from table 2 remained, with the rapid and nonprogressors being in the middle, the moderate and nonprogressors being on the bottom (despite having a lower minimum difference than the rapid and nonprogressors), and the rapid and moderate progressors having the highest maximum difference.
| Group(s) | Visit | Min Difference | Max Difference |
|---|---|---|---|
| Rapid | 1 | 39 | 52 |
| Rapid | Mid | 41 | 52 |
| Moderate | 1 | 14 | 32 |
| Moderate | Mid | 12 | 32 |
| Nonprogressor | 1 | 22 | 44 |
| Nonprogressor | Mid | 22 | 45 |
| Rapid and Moderate | 1 | 19 | 56 |
| Rapid and Moderate | Mid | 17 | 54 |
| Rapid and Nonprogressor | 1 | 24 | 54 |
| Rapid and Nonprogressor | Mid | 23 | 52 |
| Moderate and Nonprogressor | 1 | 21 | 48 |
| Moderate and Nonprogressor | Mid | 19 | 46 |
| All | 1 | 21 | 52 |
| All | Mid | 20 | 52 |
- Table 5: Comparison of all the min and max differences from tables 1, 2, 3, and 4.
Conclusions
After testing, I have come to the conclusion that my hypothesis, stated here, was false. The moderate and nonprogressors, for the most part, shared the highest amount of genetic similarity within their own groups. The rapid progressors, however, had a high level of difference within their own group, and were overall much more similar to other groups than to their own. That being said, they often had a lower maximum difference within their own group than when compared to the other groups, though the minimum difference was overall much higher within its own group then when compared to the others (15-22 higher within the rapid progressors). This indicates that the rapid progressors were uniformly different from one another. Unusually, subject 14, one of the moderate progressors, and subject 15, one of the rapid progressors, were actually very similar when viewed on the trees (Fig. 11 and Fig. 31) in both visits, which was unexpected. While they were slightly more different on the middle visit, overall they shared much more similarity than subject 15 did to either of the other rapid progressors, and than subject 14 did to either of the other moderate progressors. This may indicate that they were infected with a similar strain, but does not give any answers as to why subject 14 was only a moderate progressor and subject 15 was a rapid progressor. Unusually, the least amount of difference was actually seen in the moderate progressors as opposed to the nonprogressors, which may have been expected. The moderate progressors actually decreased in minimum difference at the middle visit, while the nonprogressors increased by one in maximum difference at the middle visit. The rapid progressors had a higher minimum difference at their midvisit than at their first visit. Overall, it can be concluded that while genetic similarity may play a role in the difference between progressor groups, it will take a much larger sample to be able to come to any distinct conclusions about the behavior of the virus in each of the progressor groups. At the moment, with the data possessed, it seems that it is relatively random whether an HIV-infected person will become a rapid progressor or not, though there may be a similarity between the viruses of the moderate progressors that could be studied further. Also, it seems that the progression of the virus doesn't change from the time of infection to the point two years following infection. A larger data sample with a greater number of timepoints would be necessary to be able to reach any further conclusions.
In regards to other papers, it doesn't appear that any of them actively studied anything similar to what I had examined in this study.
Links
Nicole Anguiano
BIOL 368, Fall 2014
Assignment Links
- Week 1 Assignment
- Week 2 Assignment
- Week 3 Assignment
- Week 4 Assignment
- Week 5 Assignment
- Week 6 Assignment
- Week 7 Assignment
- Week 8 Assignment
- Week 9 Assignment
- Week 10 Assignment
- Week 11 Assignment
- Week 12 Assignment
- Week 13 Assignment
- Week 15 Assignment
Individual Journals
- Individual Journal Week 2
- Individual Journal Week 3
- Individual Journal Week 4
- Individual Journal Week 5
- Individual Journal Week 6
- Individual Journal Week 7
- Individual Journal Week 8
- Individual Journal Week 9
- Individual Journal Week 10
- Individual Journal Week 11
- Individual Journal Week 12
- Individual Journal Week 13
- Individual Journal Week 15