BIOL368/F14:Nicole Anguiano Week 5: Difference between revisions
From OpenWetWare
Jump to navigationJump to search
(→Electronic Lab Notebook: fixed table end) |
(→Methods and Results: Added table 3 and headings) |
||
| (2 intermediate revisions by the same user not shown) | |||
| Line 30: | Line 30: | ||
===Methods and Results=== | ===Methods and Results=== | ||
*First, I compiled the sequences of each of the clones that I'd specified [[BIOL368/F14:Nicole_Anguiano_Week_4 | last week]] (also listed above) from the [http://bioquest.org/bedrock/problem_spaces/hiv/nucleotide_sequences.php bioquest web site]. I went to the [http://workbench.sdsc.edu/ Biology workbench] and created a new session, called "Research". In this session, I uploaded the file I just created with all of the sequences. | *First, I compiled the sequences of each of the clones that I'd specified [[BIOL368/F14:Nicole_Anguiano_Week_4 | last week]] (also listed above) from the [http://bioquest.org/bedrock/problem_spaces/hiv/nucleotide_sequences.php bioquest web site]. I went to the [http://workbench.sdsc.edu/ Biology workbench] and created a new session, called "Research". In this session, I uploaded the file I just created with all of the sequences. | ||
=====Visit 1===== | |||
*I began by running a CLUSTALW on each of the first visit clones. I ran one on the rapid progressors, then the moderate progressors, then the nonprogressors, importing the alignments each time so that further analysis could be performed on each alignment through CLUSTALDIST. | *I began by running a CLUSTALW on each of the first visit clones. I ran one on the rapid progressors, then the moderate progressors, then the nonprogressors, importing the alignments each time so that further analysis could be performed on each alignment through CLUSTALDIST. | ||
<!--Many thanks to http://en.wikipedia.org/wiki/Wikipedia:Picture_tutorial for help on the image formatting.--> | <!--Many thanks to http://en.wikipedia.org/wiki/Wikipedia:Picture_tutorial for help on the image formatting.--> | ||
| Line 41: | Line 43: | ||
[[Image:NP_V1_T.png|thumb|none|upright=2|alt=Nonprogressor Visit 1 Tree|<b>Figure 6</b>:The tree for subjects 2, 11, and 12 (the nonprogressors), generated by CLUSTALW.]] | [[Image:NP_V1_T.png|thumb|none|upright=2|alt=Nonprogressor Visit 1 Tree|<b>Figure 6</b>:The tree for subjects 2, 11, and 12 (the nonprogressors), generated by CLUSTALW.]] | ||
<!--*Details on what is being seen in nons here...--> | <!--*Details on what is being seen in nons here...--> | ||
*After running the CLUSTALW, I ran a CLUSTALDIST on each group to find the clustal distance matrix, and calculate the theta, min difference, and max difference (Table 1). | *After running the CLUSTALW, I ran a CLUSTALDIST on each group to find the clustal distance matrix, and calculate the theta, min difference, and max difference (Table 1). | ||
[[Image:RP_V1_CDM.png|thumb|none|500px|alt=Rapid progressor visit 1 clustal distance matrix|<b>Figure 7</b>: The clustal distance matrix for subjects 3, 11, and 15 (the rapid progressors), generated by CLUSTALDIST. The min and max difference in Table 1 was calculated by comparing across subjects and seeing what the highest and lowest values were, then multiplying by 291.]] | [[Image:RP_V1_CDM.png|thumb|none|500px|alt=Rapid progressor visit 1 clustal distance matrix|<b>Figure 7</b>: The clustal distance matrix for subjects 3, 11, and 15 (the rapid progressors), generated by CLUSTALDIST. The min and max difference in Table 1 was calculated by comparing across subjects and seeing what the highest and lowest values were, then multiplying by 291.]] | ||
| Line 88: | Line 91: | ||
| Moderate and Nonprogressor || 21 || 48 | | Moderate and Nonprogressor || 21 || 48 | ||
|- | |- | ||
| All || | | All || 21 || 52 | ||
|} | |} | ||
=====Mid-Visit===== | |||
*I moved from the first visit clones to the mid-visit clones, and ran a CLUSTALW on each of the mid-visit clones. I ran one on the rapid progressors, then the moderate progressors, then the nonprogressors, importing the alignments each time so that further analysis could be performed on each alignment through CLUSTALDIST. | |||
[[Image:RP_MV_SA.png|thumb|none|upright=3|alt=Rapid Progressor Mid-visit Sequence Alignment|<b>Figure 21</b>: The sequence alignment for subjects 3, 11, and 15 (the rapid progressors), generated by CLUSTALW. The S value in Table 3 was calculated by counting how many places there was a difference between the sequences.]] | |||
[[Image:RP_MV_T.png|thumb|none|upright=2|alt=Rapid Progressor Mid-visit Tree |<b>Figure 22</b>2: The tree for subjects 3, 11, and 15 (the rapid progressors), generated by CLUSTALW.]] | |||
<!--*Details on what is being seen in rapids here...--> | |||
[[Image:MP_MV_SA.png|thumb|none|upright=3|alt=Moderate Progressor Mid-visit Sequence Alignment|<b>Figure 23</b>: The sequence alignment for subjects 6, 8, 14 (the moderate progressors), generated by CLUSTALW. The S value in Table 3 was calculated by counting how many places there was a difference between the sequences.]] | |||
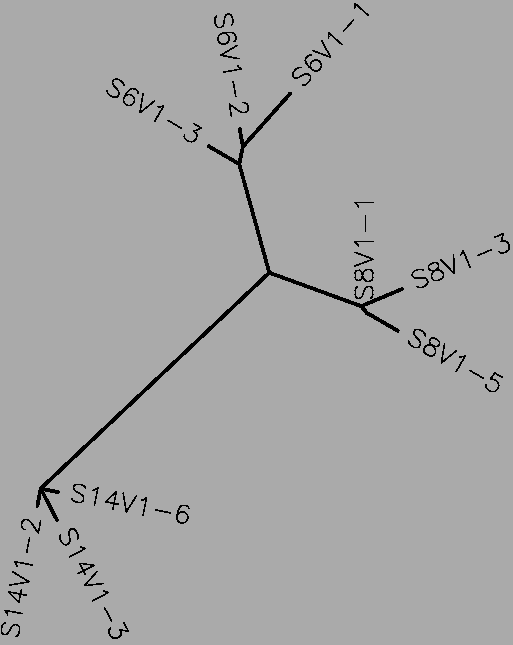
[[Image:MP_MV_T.png|thumb|none|upright=2|alt=Moderate Progressor Mid-visit Tree |<b>Figure 24</b>2: The tree for subjects 6, 8, 14 (the moderate progressors), generated by CLUSTALW.]] | |||
<!--*Details on what is being seen in moderates here...--> | |||
[[Image:NP_MV_SA.png|thumb|none|upright=3|alt=Nonprogressor Mid-visit Sequence Alignment|<b>Figure 25</b>: The sequence alignment for subjects 2, 12, 13 (the nonprogressors), generated by CLUSTALW. The S value in Table 3 was calculated by counting how many places there was a difference between the sequences.]] | |||
[[Image:NP_MV_T.png|thumb|none|upright=2|alt=Nonprogressor Mid-visit Tree |<b>Figure 26</b>2: The tree for subjects 2, 12, 13 (the nonprogressors), generated by CLUSTALW.]] | |||
<!--*Details on what is being seen in moderates here...--> | |||
*After running the CLUSTALW, I ran a CLUSTALDIST on each group to find the clustal distance matrix, and calculate the theta, min difference, and max difference (Table 3). | |||
[[Image:RP_MV_CDM.png|thumb|none|500px|alt=Rapid progressor mid-visit clustal distance matrix|<b>Figure 27</b>: The clustal distance matrix for subjects 3, 11, and 15 (the rapid progressors), generated by CLUSTALDIST. The min and max difference in Table 3 was calculated by comparing across subjects and seeing what the highest and lowest values were, then multiplying by 291.]] | |||
<!--*Details on what is being seen in rapids here...--> | |||
[[Image:MP_MV_CDM.png|thumb|none|500px|alt=Moderate progressor mid-visit clustal distance matrix|<b>Figure 28</b>: The clustal distance matrix for subjects 6, 8, and 14 (the moderate progressors), generated by CLUSTALDIST. The min and max difference in Table 3 was calculated by comparing across subjects and seeing what the highest and lowest values were, then multiplying by 288.]] | |||
<!--*Details on what is being seen in moderates here...--> | |||
[[Image:NP_MV_CDM.png|thumb|none|500px|alt=Nonprogressor mid-visit clustal distance matrix|<b>Figure 29</b>: The clustal distance matrix for subjects 2, 12, and 13 (the nonprogressors), generated by CLUSTALDIST. The min and max difference in Table 3 was calculated by comparing across subjects and seeing what the highest and lowest values were, then multiplying by 285.]] | |||
<!--*Details on what is being seen in nons here...--> | |||
{|border="1" | |||
|- | |||
!Group !! S !! θ !! Min Difference !! Max Difference | |||
|- | |||
| Rapid Progressors || 83 || - || 41 || 52 | |||
|- | |||
| Moderate Progressors || 50 || - || 12 || 32 | |||
|- | |||
| Nonprogressors || 58 || - || 22 || 45 | |||
|} | |||
*I repeated all of the above steps with the clones taken from the 2-year visit instead of the first visit. After that, I ran a CLUSTALW and CLUSTALDIST on all of the clones over all of the visits. | *I repeated all of the above steps with the clones taken from the 2-year visit instead of the first visit. After that, I ran a CLUSTALW and CLUSTALDIST on all of the clones over all of the visits. | ||
Revision as of 10:55, 30 September 2014
Electronic Lab Notebook
Answering the question: Do clones from a particular subject's group (rapidly progressing, moderately progressing, and nonprogressing), share any genetic similarity with one another? Are clones from subjects in the same group the most similar, or are they as dissimilar as clones from other groups? Does the amount of variation and similarity, if any, change from the time of the first visit to a visit after about 2 years of infection with the virus?
Below are the clones chosen and the subjects/visits they are from:
| Progressor Group | Subject | Visit # | Clones | Visit # | Clones |
|---|---|---|---|---|---|
| Rapid | 3 | 1 | 1, 2, 3 | 3 | 2, 7, 9 |
| Rapid | 11 | 1 | 3, 5, 7 | 3 | 3, 6, 9 |
| Rapid | 15 | 1 | 3, 6, 12 | 4 | 1, 3, 4 |
| Moderate | 6 | 1 | 1, 2, 3 | 5 | 2, 4, 8 |
| Moderate | 8 | 1 | 1, 3, 5 | 4 | 4, 5, 6 |
| Moderate | 14 | 1 | 2, 3, 6 | 5 | 1, 6, 7 |
| Nonprogressor | 2 | 1 | 3, 4, 5 | 3 | 5, 6, 7 |
| Nonprogressor | 12 | 1 | 1, 2, 3 | 4 | 2, 5, 10 |
| Nonprogressor | 13 | 1 | 1, 3, 4 | 3 | 1, 2, 4 |
Methods and Results
- First, I compiled the sequences of each of the clones that I'd specified last week (also listed above) from the bioquest web site. I went to the Biology workbench and created a new session, called "Research". In this session, I uploaded the file I just created with all of the sequences.
Visit 1
- I began by running a CLUSTALW on each of the first visit clones. I ran one on the rapid progressors, then the moderate progressors, then the nonprogressors, importing the alignments each time so that further analysis could be performed on each alignment through CLUSTALDIST.





- After running the CLUSTALW, I ran a CLUSTALDIST on each group to find the clustal distance matrix, and calculate the theta, min difference, and max difference (Table 1).



| Group | S | θ | Min Difference | Max Difference |
|---|---|---|---|---|
| Rapid Progressors | 83 | - | 39 | 52 |
| Moderate Progressors | 49 | - | 14 | 32 |
| Nonprogressors | 55 | - | 22 | 44 |
- Table 1
- After comparing each group with itself, I then compared across groups. I ran a CLUSTALW and CLUSTALDIST on the first visit clones from the three rapid progressors and the three moderate progressors, then the three rapid progressors and the three nonprogressors, then the three moderate progressors and the three nonprogressors. After the comparison, I ran a CLUSTALW and CLUSTALDIST on all of the visit 1 clones.








| Groups Being Compared | Min Difference | Max Difference |
|---|---|---|
| Rapid and Moderate | 19 | 56 |
| Rapid and Nonprogressor | 24 | 54 |
| Moderate and Nonprogressor | 21 | 48 |
| All | 21 | 52 |
Mid-Visit
- I moved from the first visit clones to the mid-visit clones, and ran a CLUSTALW on each of the mid-visit clones. I ran one on the rapid progressors, then the moderate progressors, then the nonprogressors, importing the alignments each time so that further analysis could be performed on each alignment through CLUSTALDIST.






- After running the CLUSTALW, I ran a CLUSTALDIST on each group to find the clustal distance matrix, and calculate the theta, min difference, and max difference (Table 3).



| Group | S | θ | Min Difference | Max Difference |
|---|---|---|---|---|
| Rapid Progressors | 83 | - | 41 | 52 |
| Moderate Progressors | 50 | - | 12 | 32 |
| Nonprogressors | 58 | - | 22 | 45 |
- I repeated all of the above steps with the clones taken from the 2-year visit instead of the first visit. After that, I ran a CLUSTALW and CLUSTALDIST on all of the clones over all of the visits.
Conclusions
Other notes
Links
Nicole Anguiano
BIOL 368, Fall 2014
Assignment Links
- Week 1 Assignment
- Week 2 Assignment
- Week 3 Assignment
- Week 4 Assignment
- Week 5 Assignment
- Week 6 Assignment
- Week 7 Assignment
- Week 8 Assignment
- Week 9 Assignment
- Week 10 Assignment
- Week 11 Assignment
- Week 12 Assignment
- Week 13 Assignment
- Week 15 Assignment
Individual Journals
- Individual Journal Week 2
- Individual Journal Week 3
- Individual Journal Week 4
- Individual Journal Week 5
- Individual Journal Week 6
- Individual Journal Week 7
- Individual Journal Week 8
- Individual Journal Week 9
- Individual Journal Week 10
- Individual Journal Week 11
- Individual Journal Week 12
- Individual Journal Week 13
- Individual Journal Week 15