AVM BIOL368 Week 4: Difference between revisions
From OpenWetWare
Jump to navigationJump to search
(→Conclusion: start conclusion) |
(→Data and Files: add him sequence info) |
||
| Line 50: | Line 50: | ||
==Data and Files== | ==Data and Files== | ||
*All files were saved to an external hard drive as well as on this page | *All files were saved to an external hard drive as well as on this page | ||
*[[Media:Visit_1_Subjects_1_thru_9_HIV.txt | HIV Sequences, Visit 1, Subjects 1-9]] | |||
*[[Media:Visit_1_Subjects_10_thru_15_HIV.txt | HIV Sequences, Visit 1, Subjects 10-15]] | |||
==Conclusion== | ==Conclusion== | ||
Revision as of 21:54, 25 September 2016
Purpose
- To continue with our class research and journal projects for the Markham et al. paper
- Continue to learn about sequencing and the similarities and differences
Methods and Results
Part 1
- Upload the “visit_1_S1_S9.txt” and “visit_1_S10_S15.txt” files from the BIOL368/F16 Week 4 page into the Biology Workbench
- Generate a multiple sequence alignment using ClustlW
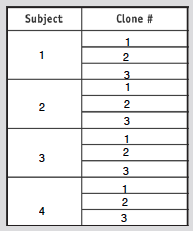
- Choose 3 clones form 4 different subjects (as shown in table below)
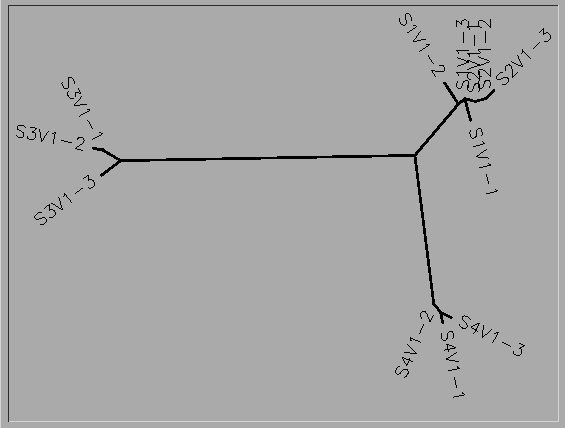
- Photo of tree shown below
Questions
- Do the clones from each subject cluster together?
- Yes
- Do some subjects’ clones show more diversity than others?
- Subject 1, strain 1 is more diverse than strains 2 and 3. Subject 2 strain 3 is more diverse than stains 1 and 2. Subject 3 strain 3 is more diverse than strains 1 and 2. Subject 4 strain 2 is more diverse than strains 1 and 3.
- Do some of the subjects cluster together?
- Yes. Subjects 1 and 2 are clustered.
- Write a brief description of the tree and how to interpret the clustering pattern with respect to the similarities and potential evolutionary relationships between subjects’ HIV sequences.
- The unrooted tree shows the similarities of the 4 subjects and clones. This tree shows that Subject 1 and 2 have very closely related viruses, and within this virus, mutations 1 and 2 of subject 2 are most closely related to mutation 3 of subject 1. All of the mutations of subject 4's virus are most closely related to each other and the mutations of subject 3 are most closely related to each other.
Part 2
- Select a sequence from last weeks assignment and align all of the clones
- Calculate S from the alignment
- S is the number of positions where there is at least one nucleotide difference across the clones
- Save the alignment for later use
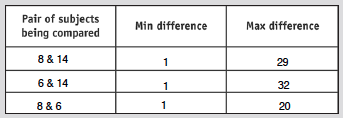
- Run an analysis for two more subjects
- Sequence differences for each subject shown at right



- (Math Is Fun was used to calculate theta)
- Within the Clustdist tool generate a distance matrix for one of your alignments
- Choose the highest and lowest pairwise scores and find the % difference
- Calculate theta for each subject
- Repeat these steps for each subject
- Table below shows results from these analysis
- Create a new alignment with all the sequences from 2 different subjects
- Use Clustdist to create a pairwise distance matrix
- Find all the min and max differences
- Results shown in table below
Data and Files
- All files were saved to an external hard drive as well as on this page
- HIV Sequences, Visit 1, Subjects 1-9
- HIV Sequences, Visit 1, Subjects 10-15
Conclusion
This exercise was done to determine if the HIV virus strains extracted from random test subjects originated from the same virus initially. After following the protocol above and creating the unrooted tree during the exercise I understood that the extracted viruses did not stem from one original viral source.
Research Project
- What is your question?
- If HIV-1 virus evolution patterns are based on virus copy number rather than CD4 T-cell decline, how would this change the subjects classification as non-progressors, moderate progressors, and rapid progress ors? Is virus copy number correlated with diversity and divergence?
- Make a prediction (hypothesis) about the answer to your question before you begin your analysis.
- There would be a shift in the subjects being grouped as moderate and rapid progressors. There is an overlap in virus number copies in Table 1 for subjects in both of these groups.
- Which subjects, visits, and clones will you use to answer your question?
- We could look at subjects 1, 3, 6 and 7 as they have very similar virus number copies. We could also choose subjects 12 and 13 to compare to those which have significantly lower virus number copies.
- You should choose a combination of subjects, visits, and clones that will add up to approximately 50 sequences. You will need about that many sequences to answer a reasonably complex question. #*However, you cannot use more because the multiple sequence alignment tool cannot handle more than that many sequences.
Justify why you chose the subjects, visits, and clones you did.
Acknowledgments
- Worked in collaboration with Courtney L. Merriam in class.
- Template followed from week 4 assignment BIOL368/F16
- Help from our professor, Dr. Dahlquist in class.
- Note: While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source.
Avery Vernon-Moore 14:42, 21 September 2016 (EDT)
References
Weekly Assignments:
Assignment 1 Assignment 2 Assignment 3 Assignment 4 Assignment 5 Assignment 6 Assignment 7 Assignment 8 Assignment 9 Assignment 10 Assignment 11 Assignment 12 Assignment 14 Assignment 15
Individual Journals:
"Week 1: Create Wiki Page" Individual Journal 2 Individual Journal 3 Individual Journal 4 Individual Journal 5 Individual Journal 6 Individual Journal 7 Individual Journal 8 Individual Journal 9 Individual Journal 10 Individual Journal 11 Individual Journal 12 No Week 13 Assignment Individual Journal 14 Individual Journal 15
Class Journal 1 Class Journal 2 Class Journal 3 Class Journal 4 Class Journal 5 Class Journal 6 Class Journal 7 Class Journal 8 Class Journal 9 Class Journal 10 Class Journal 11 Class Journal 12 Class Journal 14 Class Journal 15